Eukarya
General Introduction
Lists
Organization
Origins of the Eukarya
Introduction
Cell Membranes and Walls
Cytoskeleton
General Metabolism
Internal Membranes
Chromosome and Genome
Abrupt Concluding Remarks

| Eukarya | ||
| Eukarya | Eukarya origins - 1 |
| Life | Time |
| Abbreviated Dendrogram Life
├─Eubacteria
└─┬─Archaea
└─Eukarya
├─Metamonada
└─┬─Discicristata
└─┬─Rhizaria
└─┬─┬─┬─Alveolata
│ │ └─Chromista
│ └─Plantae
└─Stem Metazoa
├─Fungi
└─Metazoa
|
Contents
Eukarya  |
 This is likely to become our longest and most ambitious discussion yet. We're going to talk here about where the eukaryotes came from and what makes them different -- and they are superficially quite different from the other domains. In order to set the stage, we need to get a couple of things straight with the (hypothetical) reader. First, and because the bacteria and Eukarya are very different from each other, we need to understand that they are not so different that we should think of them as only remotely related to each other. Second, we're going to need to have a frank discussion about our approach to this topic.
This is likely to become our longest and most ambitious discussion yet. We're going to talk here about where the eukaryotes came from and what makes them different -- and they are superficially quite different from the other domains. In order to set the stage, we need to get a couple of things straight with the (hypothetical) reader. First, and because the bacteria and Eukarya are very different from each other, we need to understand that they are not so different that we should think of them as only remotely related to each other. Second, we're going to need to have a frank discussion about our approach to this topic.
In spite of the differences in structure and chemistry between bacteria and Eukarya, bear in mind that both work more or less the same way. All organisms descended from LUCA use the same basic inventory of amino acids and nucleotides to make proteins and nucleic acids, respectively. With exceptions, living species share a large number of common sugar monomers and, with even more exceptions, common lipids. The predominant amino acids in bacteria and eukaryotes are all left-handed, and the sugar monomers are right-handed. Proteins in both bacteria and eukaryotes are coded by DNA, transcribed into RNA and translated into protein, using essentially identical genetic codes, and biochemical procedures which have numerous similarities. The usual carrier of short-term energy is  ATP in all living organisms, the electron carriers always include NAD and its close relatives. Some of the metabolic intermediates used to generate energy are almost universal. Perhaps most significantly, a good many protein families have specific, identifiable homologues in every organism alive today.
ATP in all living organisms, the electron carriers always include NAD and its close relatives. Some of the metabolic intermediates used to generate energy are almost universal. Perhaps most significantly, a good many protein families have specific, identifiable homologues in every organism alive today.
The point here is that, considering the entire universe of possible biochemistries, all extant life forms are fairly similar, similarly specialized, and thus closely related. We will be emphasizing those similarities in order to get a handle on the evolutionary course of the differences.
Our approach to this enormous topic has, itself, evolved. The original idea was to drag in the usual suspects: Cavalier-Smith, Martin, Woese, Gupta -- you know the lot we're referring to. Then we'd describe the basic outlines of their various theories, do a little stylish arm-waving and be done. For a more succinct treatment of origins of Eukarya, visit the "Endosymbiosis - The Origin of the Eukaryotes" page of the Virtual Fossil Museum.
That scheme fell apart in short order. The problem is entirely due to our own poor attitude and bad habits. Nostra maxima culpa. Here is a list of our failings, in the form of credo:
 1) We think that normal, Darwinian evolution is the most parsimonious default explanation in almost every case. Consequently, we are unimpressed with theories invoking weird chimaeras and massive horizontal gene transfer ("HGT") where there are more prosaic explanations. But see, for example, Simonson et al. (2005), Baluška et al. (2004), Martin & Russell (2002), Woese (2002), Margulis et al. (2000), López-García & Moreira (1999), Gupta & Golding (1996) -- and we even get a little suspicious of phrases like "quantum evolution." Cavalier-Smith 2002, 2002a, 2006). Unless (a) the normal rules of evolution absolutely can't explain the observed trait distribution and/or (b) there is some other very good reason to think that the rules have been bent (as in chloroplasts), we ought to prefer speciation to special pleading. Thus far, we have not seen a case which seems to satisfy either of these conditions.
1) We think that normal, Darwinian evolution is the most parsimonious default explanation in almost every case. Consequently, we are unimpressed with theories invoking weird chimaeras and massive horizontal gene transfer ("HGT") where there are more prosaic explanations. But see, for example, Simonson et al. (2005), Baluška et al. (2004), Martin & Russell (2002), Woese (2002), Margulis et al. (2000), López-García & Moreira (1999), Gupta & Golding (1996) -- and we even get a little suspicious of phrases like "quantum evolution." Cavalier-Smith 2002, 2002a, 2006). Unless (a) the normal rules of evolution absolutely can't explain the observed trait distribution and/or (b) there is some other very good reason to think that the rules have been bent (as in chloroplasts), we ought to prefer speciation to special pleading. Thus far, we have not seen a case which seems to satisfy either of these conditions.
2) We ought to look much harder to find the plesiomorphic ("primitive") state. We know relatively little of the diversity of either protists or bacteria -- even the ones that are alive today. Cavalier-Smith 2004), Fieseler et al. (2004), Moreira & Lópex-García (2002); López-García et al. (2001); Roger (1999). As we will see, the perceived need to uncork the genie of HGT is far less than commonly believed -- if we really look hard enough at the cast of characters we already have. The gaps between Archaea, Eukarya, and bacteria are no greater than the gaps between fishes and Tetrapoda, or reptiles and Mammalia, appeared to be a century ago. Then, as now, what's needed is hard work and detailed observation. Airy theorizing (of the type we usually do on Palaeos, for example) isn't going to get us there.
3) We have to be very careful about making generalizations. [1] Particularly when we're looking at things that happened a couple of billion years ago, we can get into trouble very quickly ignoring oddball exceptions. Take, for example, the DNA of Eubacteria. The bacterial chromosome is almost always characterized as small, circular, uncondensed, haploid, and histone-free. All those adjectives apply to the genomes of many Eubacteria, and some apply to just about
 all the Eubacteria. But, once Bendich & Drlica (2000) really started looking, "[W]e found so many exceptions to commonly held views about chromosome multiplicity, ploidy, linearity, heterochromatinization, partitioning, and histone-based DNA packaging that we were forced to conclude that chromosomal properties do not correlate well with the presence or absence of a nuclear membrane." For that matter, some bacteria even have the nuclear membrane. Fuerst (2004).
all the Eubacteria. But, once Bendich & Drlica (2000) really started looking, "[W]e found so many exceptions to commonly held views about chromosome multiplicity, ploidy, linearity, heterochromatinization, partitioning, and histone-based DNA packaging that we were forced to conclude that chromosomal properties do not correlate well with the presence or absence of a nuclear membrane." For that matter, some bacteria even have the nuclear membrane. Fuerst (2004).
4) A billion years is a hell of a long time. Maybe that only means that the weird chimaeras of various popular models might have occurred. In theory, it could happen. Remember The Island of Dr. Moreau? But our prejudice is to the contrary. That is, the longer the time interval, the more likely that something evolved in the usual, boring, incremental, non-cinematic way. After all, if some hapless victim walks into Dr. Moreau's laboratory and comes out again in a couple of days looking like a wart hog, we're going to suspect that something other than natural selection was at work. On the other hand, if the interval is a couple of gigayears, we are more inclined to attribute the transformation to stepwise evolution and a particularly unfortunate spasm of homoplasy.
5) Finally, and as always, we look upon sequence-based phylogenies with mild distaste. As a matter of fact, while doing the research for this essay, we discovered, with no small delight, that at least some of the biological community seem to be getting the message. We will discuss a number of recent papers in which respectable scientists likewise found that structure is a far better key to phylogeny than sequence. For once, we can smugly claim that "we told you so."
So, after assiduous application of our various prejudices and groundless assumptions, what's left? Initially, we concluded that we were left with Thomas Cavalier-Smith. Prof. Cavalier-Smith of Oxford University has produced a large body of work which is well-regarded. Still, he is controversial in a way that is a bit difficult to describe. The issue may be one of writing style. Cavalier-Smith has a tendency to make pronouncements where others would use declarative sentences, to use declarative sentences where others would express an opinion, and to express opinions where angels would fear to tread. In addition, he can sound arrogant, reactionary, and even perverse. On the other, he has a long history of being right when everyone else was wrong. To our way of thinking, all of this is overshadowed by one incomparable virtue: the fact that he will grapple with the details. This makes  for very long, very complex papers and causes all manner of dark murmuring, tearing of hair, and gnashing of teeth among those tasked with trying to explain his views of early life. See, e.g., Zravý 2001); Patterson 1999). Nevertheless, he deals with all of the relevant facts.
for very long, very complex papers and causes all manner of dark murmuring, tearing of hair, and gnashing of teeth among those tasked with trying to explain his views of early life. See, e.g., Zravý 2001); Patterson 1999). Nevertheless, he deals with all of the relevant facts.
Thus, as Plan B, we determined to outline some of Cavalier-Smith's views. Hard work, but the project was appealing because it didn't require much original thought on our part. Unfortunately, this second iteration didn't work any better than the first. Cavalier-Smith has reached the conclusion that the Archaea and Eukarya are sister groups. He calls the crown group of Archaea + Eukarya = Neomura. No problem so far. That's probably the majority view. However, Cavalier-Smith also asserts that the Eubacteria are paraphyletic. That is, he argues that LUCA was a perfectly ordinary bacterium -- perhaps living at the split between green sulfur and non-sulfur bacteria. Cavalier-Smith (2006). Much later, maybe less than 1000 Ma ago, some more derived bacterium became the first neomuran, the last common ancestor of all eukaryotes and archaeans. This opinion is heterodox but, except for the timing, his arguments seemed quite persuasive. Alternatively, perhaps it was late and we were simply tired of (mentally) arguing with him. Either way, we were willing to accept the whole mess if it meant we could avoid doing any real work.
The dicey part was that, as we closed in on the end of Cavalier-Smith's story, we found that we absolutely couldn't accept his nominee for the ancestral neomuran. Cavalier-Smith argues that the first neomuran was not just any old bacterium but, specifically, a moderately derived Gram-positive bug and probably a member of the Actinobacteria. We reluctantly concluded that an equally good, and perhaps better argument, can be made that the first neomuran was, instead, a moderately basal Gram-negative bacterium. Granted, this is a little like getting to the end of an Agatha Christie and deciding that Hercule Poirot had misidentified the killer -- or possibly even worse, since lots of people feel that way about The Murder of Roger Ackroyd. But there it is, and, as a result, we really will have to cover the ground in detail. We will omit some issues (e.g. introns), and breeze over most others, for lack of time, space, and patience. Other important matters, such as the position of the root within Eukarya, aren't all that relevant to our issue. That issue is: assuming Cavalier-Smith is generally correct about bacterial phylogeny, where did the Eukarya come from?
| Root
├─Eukarya
├─Archaea
│ ├─Euryarcheota
│ └─Crenarcheota
└──┬─Chlamydiales
└─┬─Spirochaetes
└─┬─┬─┬─Aquifex
│ │ └─Thermotoga
│ └─Proteobacteria
└─┬──┬─Cyanobacteria
│ └─┬─Deinococcus
│ └─Actinobacteria
└─Endobacteria |
Root
├─Deinococcus
└─┬─Cyanobacteria
└─┬─┬─┬─Aquifex
│ │ └─Thermotoga
│ └─┬─Spirochaetes
│ └─┬─Chlamydiales
│ └─Proteobacteria
└─┬─Endobacteria
└─┬─Actinobacteria
└─┬─Eukarya
└─Archaea
├─Euryarcheota
└─Crenarcheota |
The very broadest outlines of bacterial phylogeny are beginning to settle down. It often doesn't look that way, because the both the location of LUCA and the branch point of Neomura are disputed. So, for example, a simplified version of the supertree of Daubin et al. (2001) is shown in the figure, compared to the general scheme of Cavalier-Smith (2006). The two trees have very significant differences, and entirely different roots, but the main groupings, and even many of the specific linkages, are preserved.
In essence, we're dealing with four high-level groups. Maybe they're clades. Maybe not. The various members of this quartet are as follows. Phylogenetically defined taxa are in bold, but mostly we'll simply refer to these groups by their parts in the bacterial chorus and avoid worrying about the vagaries of nomenclature:
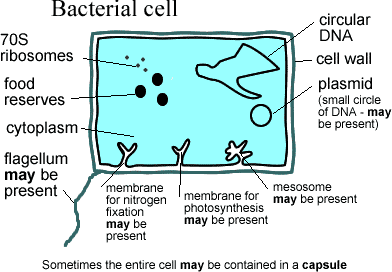
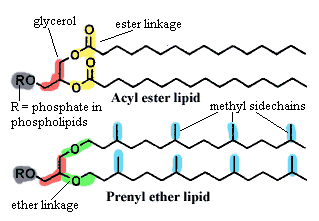
Sopranos: the Neomura, Eukarya + Archaea. Classically, the Eukarya include everything with a nucleus. The Archaea are a varied lot of bacteria -- mostly thermophiles. They have a number of characteristic features, the most easily remembered of which is a cell membrane composed of prenyl ether lipids, but very few actual synapomorphies (Cavalier-Smith, 2006). They have DNA-associated enzymes which are relatively similar to those of Eukarya.
Altos: the Gram Positives or Unibacteria. For our
 purposes, Bacillus > E.coli. Unlike most bacteria, the Gram Positives have no outer membrane external to the cell wall. They are divided into (a) the Actinobacteria, roughly equivalent to the High G+C group (perhaps Streptomyces > Bacillus) which is frequently filamentous and (importantly) possesses a 20S proteasome; and (b) the Endobacteria or low-G+C group, a probably paraphyletic group with all the other Gram positives.
purposes, Bacillus > E.coli. Unlike most bacteria, the Gram Positives have no outer membrane external to the cell wall. They are divided into (a) the Actinobacteria, roughly equivalent to the High G+C group (perhaps Streptomyces > Bacillus) which is frequently filamentous and (importantly) possesses a 20S proteasome; and (b) the Endobacteria or low-G+C group, a probably paraphyletic group with all the other Gram positives.
Tenors: the Firmicutes, defined for our purposes as E. coli > Bacillus. These are conventional Gram-negative bacteria. Their most noted members are the Proteobacteria. Typically they have a second membrane outside the cell wall, which accounts for their failure to respond to Gram staining.
Basses: the extremely paraphyletic (if Cavalier-Smith is correct) Gram-negative basal bacteria, including green bacteria of all kinds. Finally, the famous duo comprised of the Thermotogales and Aquifex seem to be either very low tenor/baritones, or contrabasses. In any case, they're very odd and we have no explanation for them.
 In order to achieve a more harmonious arrangement, we're going to create a "baritone" group by splitting out a bunch of Firmicutes, comprising some, perhaps most, of the tenors who are basal to the Proteobacteria. This group, which is widely believed to be related (if paraphyletic), is anchored on the Planctomycetes, but also includes the Verrucomicrobia, Chlamydiaceae, probably along with the Spirochaetes and the newly-discovered Poribacteria. Strous et al. 2006); Fuerst (2004, 2005); Teeling et al. (2004); Glöckner et al. (2003). Our emphasis on this group will make the piece a little heavy on lower voices; but that was good enough for Mozart (Le Nozze di Figaro), so you'll just have to put up with it. We will introduce the Planctomycetes in particular, and talk about the other baritones as they come up in the discussion.
In order to achieve a more harmonious arrangement, we're going to create a "baritone" group by splitting out a bunch of Firmicutes, comprising some, perhaps most, of the tenors who are basal to the Proteobacteria. This group, which is widely believed to be related (if paraphyletic), is anchored on the Planctomycetes, but also includes the Verrucomicrobia, Chlamydiaceae, probably along with the Spirochaetes and the newly-discovered Poribacteria. Strous et al. 2006); Fuerst (2004, 2005); Teeling et al. (2004); Glöckner et al. (2003). Our emphasis on this group will make the piece a little heavy on lower voices; but that was good enough for Mozart (Le Nozze di Figaro), so you'll just have to put up with it. We will introduce the Planctomycetes in particular, and talk about the other baritones as they come up in the discussion.
The Planctomycetes were almost completely ignored until about ten years ago. Two factors have changed that. First, they have come into their own in a practical way because they can degrade wastewater and sludge as almost nothing else can. This is probably because the Planctomycetes can do things with nitrogen which no other bacteria can manage. This includes the degradation of chitin, which eukaryotes create in prodigious quantities and which few organisms can digest. Second, they might represent a phylogenetic link with the Neomura -- but even if they do not, they offer a series of important lessons in what bacteria are capable of producing by normal, Darwinian evolution. Fuerst (2004).
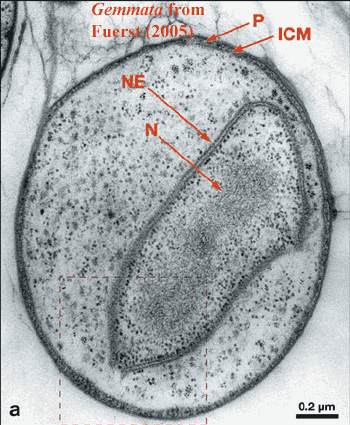
The well-characterized Planctomycete genera include Pirellula, Rhodopirellula (formerly known as Pirellula sp. strain 1), Planctomyces, Isosphaera, and Gemmata. These genera are all facultatively aerobic. Phylogenetically basal to these forms are a group of anaerobic chemoautotrphs who metabolize ammonia and nitrates anaerobically to form molecular nitrogen gas, i.e., the "anammox" pathway. Fuerst (2005). These genera have never been cultured and are thus often referred to as, e.g. "Candidatus" Brocadia anammoxidans. We will omit such formalities. However, it is worth remembering that -- absent a pure culture -- there is a slight chance that any biochemical result may be an artifact of contamination.
 In the last decade it has become obvious that the planctomycetes are some of the most diverse and ubiquitous organisms on Earth: "They have been found to be abundant in various habitats including terrestrial and aquatic habitats differing in salinity (from hypersaline to freshwater), oxygen availability (from the oxic water-column to anoxic sediments), trophic level (from oligotrophic lakes to eutrophic wastewater) and temperature (from cold-water marine snow to hot springs) ... Planctomycetes have even been isolated from the digestive tracts of crustaceans." Teeling et al. (2004); see also Brochier (2002). Their importance in the detrital marine "snow" is particularly significant, since this represents one of the main sources of carbon burial in the planetary carbon cycle. Fuerst (1995).
In the last decade it has become obvious that the planctomycetes are some of the most diverse and ubiquitous organisms on Earth: "They have been found to be abundant in various habitats including terrestrial and aquatic habitats differing in salinity (from hypersaline to freshwater), oxygen availability (from the oxic water-column to anoxic sediments), trophic level (from oligotrophic lakes to eutrophic wastewater) and temperature (from cold-water marine snow to hot springs) ... Planctomycetes have even been isolated from the digestive tracts of crustaceans." Teeling et al. (2004); see also Brochier (2002). Their importance in the detrital marine "snow" is particularly significant, since this represents one of the main sources of carbon burial in the planetary carbon cycle. Fuerst (1995).
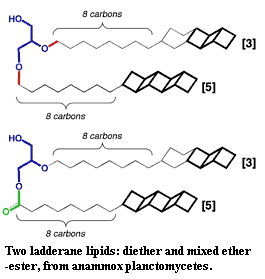
Structurally and biochemically, the baritones generally, and the planctomycetes in particular, have any number of supposed neomuran features scattered among their members. The most famous of these (to the extent that anything about the Planctomycetes can be described as "famous") is probably the true folded double-membrane nuclear compartment in Gematta obscuriglobus. In addition, some of the planctomycetes confound the acyl ether/prenyl ether dichotomy by manufacturing "ladderane" lipids with both types of linkage, in addition to unique strings of fused cyclobutane rings.
But we will not plunge headlong into the bizarre world of the Planctomycetes. Instead, we will disclose the details as they come up in a more systematic discussion of the eukaryotic cell and its evolution, to which we now turn.
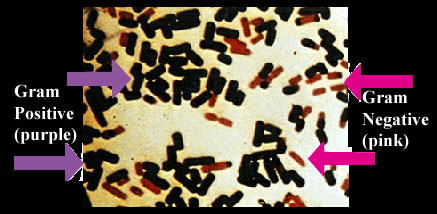 As usual with single-celled organisms, we start from the outside and work inwards. Eukaryotes lack an outer membrane. The cell is bounded by a single membrane. Gram negative bacteria are typically "negative" because they have an outer membrane, outside the cell wall. This is one of the reasons why Cavalier-Smith is convinced that the ancestor of Neomura had to be an alto, a Gram-positive organism. He makes a strong, scenario-based argument that the outer membrane has great evolutionary stability and was only lost once, in the Altos, and that the Sopranos inherited this trait directly. Cavalier-Smith 2006).
As usual with single-celled organisms, we start from the outside and work inwards. Eukaryotes lack an outer membrane. The cell is bounded by a single membrane. Gram negative bacteria are typically "negative" because they have an outer membrane, outside the cell wall. This is one of the reasons why Cavalier-Smith is convinced that the ancestor of Neomura had to be an alto, a Gram-positive organism. He makes a strong, scenario-based argument that the outer membrane has great evolutionary stability and was only lost once, in the Altos, and that the Sopranos inherited this trait directly. Cavalier-Smith 2006).
The difficulty with this proposition is that it is either false or irrelevant. First, the Planctomycetes also lack an outer membrane -- we think. The fraternity of microbiologists who publish on the morphology of the Planctomyces has been curiously reticent about this. They do not commit themselves on this issue, or even mention it. Their studied indifference is, most likely, the result of an entirely understandable unwillingness to take on issues of homology. We'll dodge that one ourselves, since it isn't the issue. Cavalier-Smith's assertion is that an actual outer membrane was lost only once. The Planctomycetes may or may not have something homologous, but they have no outer membrane and even lack the genes to make some of the essential lipopolysaccharide linkages. Glöckner et al. (2003).
Second, it is relatively easy to create "L-forms" of various Gram-negative bacteria. These are strains selected by growth in a medium in which cell walls cannot be maintained. After extensive selection, stable variants are produced which do not produce a cell wall even under permissive conditions. The relevant point is that some of these strains also lose the outer membrane. Onoda et al. 2000); Dienes & Bullivant (1968). Thus, the loss of the outer membrane cannot be as rare or traumatic as Cavalier-Smith makes out.[2]