Origins of the Eukarya - 4
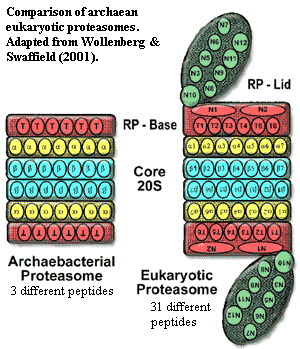 We have briefly discussed proteasomes elsewhere. This is the second of the rough spots to get over. As noted in the glossary entry at the link, proteasomes are related to chaperonins, such as GroEl. However, the function of the proteasome is to destroy proteins, rather than to rehabilitate them. For the Planctomycetes, this this is not only a rough spot. It may be the end of the road.
We have briefly discussed proteasomes elsewhere. This is the second of the rough spots to get over. As noted in the glossary entry at the link, proteasomes are related to chaperonins, such as GroEl. However, the function of the proteasome is to destroy proteins, rather than to rehabilitate them. For the Planctomycetes, this this is not only a rough spot. It may be the end of the road.
Cavalier-Smith (e.g., 2006) makes the important and powerful observation that only Actinobacteria and neomurans share the basic core 20S proteasome structure. In fact, the case has recently become even stronger, with the completed crystal structure of the Mycobacterium proteasome. Amoils (2006) (research summary). One of the differences between actinobacterial and eukaryotic proteasomes was thought to be that the latter were "closed" at the two ends of the barrel and required (at least) an RP base unit for activity. However, the structure of the 20S proteasome from Mycobacterium shows that it, too, is a closed barrel and must associate with other proteins to accept substrate peptides for destruction in its interior, thus increasing the similarity between the actinobacterial and neomuran proteasomes.
The proteases making up the 20S proteasome are, to be sure, AAA+ proteases -- an enzyme family found in all life. Wollenberg & Swaffield (2001). Yet, unaccountably, most AAA+ proteases stubbornly refuse to spontaneously self-assemble into αββα-stacked heptameric rings.
 As Cavalier-Smith (2002, 2002a) quite correctly points out, the further elaboration of the proteasome was a key development in the evolution of eukaryotes. Eukaryotes have an elaborate cascade of controls which attach one or several copies of a small protein, ubiquitin, as a "tag" onto other proteins. The additional controls represented by the eukaryotic "lid" on the proteasome recognize and preferentially digest multiply-tagged proteins. The ubiquitin cascade operates (among other things) to tag and remove proteins specific for particular phases of the mitotic cycle. Thus, the development of this system was probably critical to the evolution of mitosis in eukaryotes. For reviews, see Myung et al. (2001), Glickman & Ciechanover (2002). Ubiquitin itself is derived from sulfur-transferring enzymes involved in the synthesis of enzymatic co-factors molybdopterin, thiamin). These are widely distributed in all groups of bacteria and Archaea and don't seem to have any useful phylogenetic signal. Xu et al. (2006); Iyer et al. (2006).
As Cavalier-Smith (2002, 2002a) quite correctly points out, the further elaboration of the proteasome was a key development in the evolution of eukaryotes. Eukaryotes have an elaborate cascade of controls which attach one or several copies of a small protein, ubiquitin, as a "tag" onto other proteins. The additional controls represented by the eukaryotic "lid" on the proteasome recognize and preferentially digest multiply-tagged proteins. The ubiquitin cascade operates (among other things) to tag and remove proteins specific for particular phases of the mitotic cycle. Thus, the development of this system was probably critical to the evolution of mitosis in eukaryotes. For reviews, see Myung et al. (2001), Glickman & Ciechanover (2002). Ubiquitin itself is derived from sulfur-transferring enzymes involved in the synthesis of enzymatic co-factors molybdopterin, thiamin). These are widely distributed in all groups of bacteria and Archaea and don't seem to have any useful phylogenetic signal. Xu et al. (2006); Iyer et al. (2006).
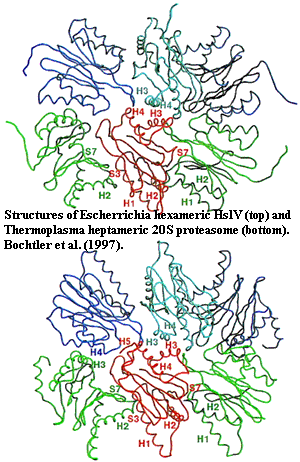 This would be rather discouraging -- and impossible to reconcile fully with the rest of the evidence -- except that we may have overstated the case. While it is true that the 20S actinobacterial proteasomes are quite similar to their neomuran counterparts, the Gram-negative bacteria have HslV complexes which are almost as similar. True, they form rings which are hexameric, rather than heptameric; but they are not really so different for all that [17]. Further, recent work has shown that the association between subunits in eukaryotic proteasomes is surprisingly flexible.
This would be rather discouraging -- and impossible to reconcile fully with the rest of the evidence -- except that we may have overstated the case. While it is true that the 20S actinobacterial proteasomes are quite similar to their neomuran counterparts, the Gram-negative bacteria have HslV complexes which are almost as similar. True, they form rings which are hexameric, rather than heptameric; but they are not really so different for all that [17]. Further, recent work has shown that the association between subunits in eukaryotic proteasomes is surprisingly flexible.
Perhaps more important than debating the subtleties of "similarity," is the phylogenetic distribution of HslV. Actinobacteria have proteasomes, rather than HslV. Eukaryotes have proteasomes in addition to HslV. Couvreur et al. (2002); Ruiz-González & Marin (2006). As usual, we can wave our arms and shout the mystic formula: "Abracadabra Horizontalgenetransfer!" Then, with a puff of smoke, everyone's genome will be magically shuffled to suit our tastes. This ranks right up there with "evil spirits," international conspiracies, and psychic auras as a labor-saving, thought-avoidance technique. However, we ought to at least consider the possibility that inheritance of HslV was vertical, which would suggest that the proteasome was invented at least twice. That might explain, among other things, why HslV is (by sequence) actually closer to the β-units of neomuran proteasomes (18-20% identity) Bochtler et al., 1997; Couvreur et al., 2002) than actinobacterial
α-units are to the α-units of Neomura (15% identity) (Gille et al., 2003).
This is a close call. The evidence clearly favors the altos here, perhaps decisively; and our desire to complete the research for this entire discussion -- never all that strong at the best of times -- nearly crumbled before the temptation of this unique, but convenient, excuse to bugger out. Unfortunately, all other evidence we turned up seems to favor the baritones, particularly in the case of the internal cell membranes, which we will consider next.
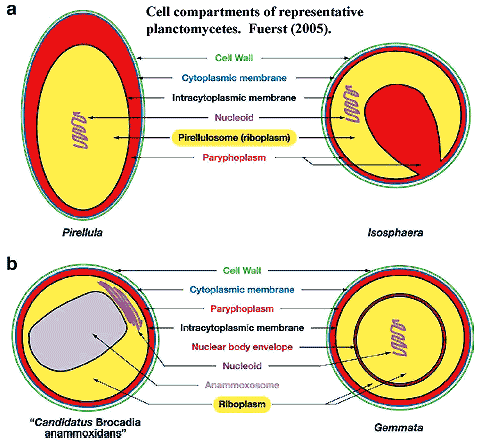 Nothing could be more obvious about the Eukarya than their internal membrane system. The nucleus, endoplasmic reticulum and Golgi apparatus make the eukaryotic cell instantly recognizable. Likewise, the internal compartmentation of planctomycetes also makes them instantly recognizable among bacteria. However, this comparison is potentially misleading in at least two ways.
Nothing could be more obvious about the Eukarya than their internal membrane system. The nucleus, endoplasmic reticulum and Golgi apparatus make the eukaryotic cell instantly recognizable. Likewise, the internal compartmentation of planctomycetes also makes them instantly recognizable among bacteria. However, this comparison is potentially misleading in at least two ways.
First, intracellular membrane systems are much more common in bacteria than is sometimes supposed. Fuerst (2005) has compiled some examples, including:
acidocalcisome-like organelle of Agrobacterium tumefaciens and Rhodospirillum rubrum; the chromatophores of purple nonsulfur photosynthetic bacteria; the chlorosomes of green sulfur photosynthetic bacteria; thylakoids of photosynthetic cyanobacteria; intracellular membranes of chemoautotrophic and methanotrophic bacteria; RuBisCO-containing carboxysomes of chemo- and photoautotrophic bacteria such as nitrifiers, sulfur-oxidizing thiobacilli, and cyanobacteria; enterosomes of Salmonella enterica; and magnetosomes of magnetotactic bacteria.
Second, we can't assume homology between these membranes and topologically similar systems in eukaryotes. Even within the Planctomyces, the arrangements vary considerably.
And now, having carefully crafted the impression of cautious and dispassionate judgment (in the approved manner for review writers), we may proceed to leap wildly about from one unsubstantiated conclusion to the next, like a jackrabbit on roller skates.
 Although internal membrane systems are common enough in bacteria, internal membranes which wall off the chromosome are not. In fact, such membranes are unique to the Planctomycetes. That's not surprising, since it creates an awkward problem. Bacteria couple transcription with translation. That is, protein is synthesized while RNA is still bound to the chromosome. How, then, does one supply the cell wall, plasma membrane and external cytoplasm with protein? Evidently, planctomycetes have some sort of post-translational transport mechanism which can carry protein through the intracytoplasmic membrane and direct it to the appropriate destination.
Although internal membrane systems are common enough in bacteria, internal membranes which wall off the chromosome are not. In fact, such membranes are unique to the Planctomycetes. That's not surprising, since it creates an awkward problem. Bacteria couple transcription with translation. That is, protein is synthesized while RNA is still bound to the chromosome. How, then, does one supply the cell wall, plasma membrane and external cytoplasm with protein? Evidently, planctomycetes have some sort of post-translational transport mechanism which can carry protein through the intracytoplasmic membrane and direct it to the appropriate destination.
To appreciate this problem, it's important to note two further items. First, the usual bacterial chromosome is actually in contact with the plasma membrane (textbook illustrations to the contrary notwithstanding). Thus membrane proteins are probably inserted as they are created. Second, it appears that the planctomycete intracytoplasmic membrane is generally not in direct contact with the plasma membrane, and no DNA or ribosomes are found outside the intracytoplasmic membrane. Lindsay et al. (2001). [15] Thus, a novel intermediate transport step must be involved.
Perhaps the vesicles which seem to show up in freeze-fracture preparations are involved in transport. See, Fuerst (2005: fig. 5), Lindsay et al. (2001: figs. 2B, 8A, 8B). However, since no one seems to have made an issue of them, this is pure speculation. Another possibility is that the original system for shuttling materials from the plasma membrane to the outer membrane has been exapted for this novel purpose. Again, there is no worthwhile evidence.
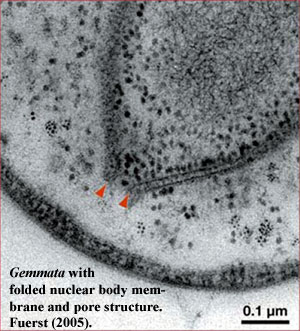
Things get yet more interesting (or simply messier) in Gemmata and closely-related species. The intracytoplasmic membrane is a single-layered membrane with no obvious direct homology to eukaryotic internal membranes. Gemmata is different. In addition to the intracytoplasmic membrane, 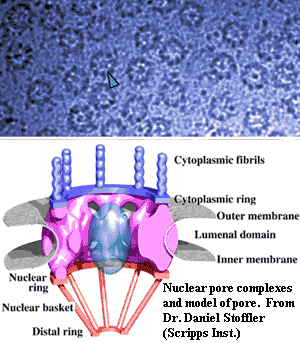 Gemmata has a double-walled membrane around the chromosome -- just like a nucleus. Like a nuclear membrane, the "nuclear body membrane" in Gemmata appears to have pores of a sort, which seem to indicate that the double membrane here, as in eukaryotes, is actually a single membrane folded back on itself, since the two layers are continuous around the pores. Fuerst (2005). Note that all of the DNA is located inside the nuclear membrane, but ribosomes are found both inside and outside. Thus, Gemmata must transport mRNA across the membrane, and at least some translation must be uncoupled from transcription -- the hallmark of the eukaryote condition.
Gemmata has a double-walled membrane around the chromosome -- just like a nucleus. Like a nuclear membrane, the "nuclear body membrane" in Gemmata appears to have pores of a sort, which seem to indicate that the double membrane here, as in eukaryotes, is actually a single membrane folded back on itself, since the two layers are continuous around the pores. Fuerst (2005). Note that all of the DNA is located inside the nuclear membrane, but ribosomes are found both inside and outside. Thus, Gemmata must transport mRNA across the membrane, and at least some translation must be uncoupled from transcription -- the hallmark of the eukaryote condition.
The presence of these nuclear-pore-like structures also provides us with a useful reality check. Not just any old hole in the membrane will do. Nuclear pores have a very specific structure and relationships. Most significantly, the lumenal domain (see figure) is dominated by pore proteins that have a rather distinctive structural motif: an N-terminal β-propeller domain, followed by an α-solenoid domain, the latter consisting of a series of α-helices separated by loops. It has recently been determined (by some inspired and beautiful in silico "experiments") that these proteins are related to the vesicle coating complexes essential for transport in the eukaryotic cell. Devos et al. (2004). Finally, the outer leaflet of the nuclear membrane is linked to, and likely continuous with, the endoplasmic reticulum in eukaryotes. Du et al. (2004). We cannot claim that the planctomycetes have all these accessories, but the data is interesting all the same.
The N-terminal β-propeller domain, then α-solenoid motif is found in planctomycetes. By a ridiculous stroke of blind luck, we happened to be reading Strous et al. (2006) at about the same time as we saw the Devos paper. Strous includes diagrams of some possible anammox-related operons likely to be expressed in the membrane of the anammoxosome. One of these is shown as having an N-terminal β-propeller sequence, followed by a possible cytochrome c peroxidase. We wondered if -- just by chance -- the crystal structure of cytochrome c peroxidase had been determined; and, sure enough, it had.
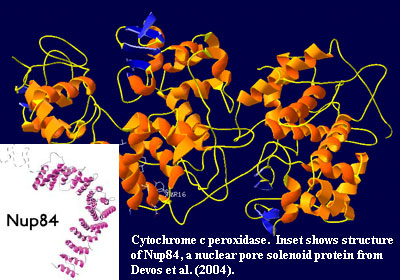 In fact, cytochrome c peroxidase turns out to a strapping example of an α-solenoid system. Even the small β domains (in blue) take on the correct antiparallel, triangular appearance of "propeller" blades when viewed from an appropriate angle. Bear in mind, however, that this is the wrong planctomycete (Keunenia, rather than Gemmata) and the wrong membrane (anammoxosome, not nuclear). [16]
In fact, cytochrome c peroxidase turns out to a strapping example of an α-solenoid system. Even the small β domains (in blue) take on the correct antiparallel, triangular appearance of "propeller" blades when viewed from an appropriate angle. Bear in mind, however, that this is the wrong planctomycete (Keunenia, rather than Gemmata) and the wrong membrane (anammoxosome, not nuclear). [16]
From an ultrastructural point of view, Gemmata's nuclear pores look more or less correct. They do not fuse with an endoplasmic reticulum, since Gemmata lacks an ER system -- sort of. Actually, Lindsay et al. (2001: figs. 7A-C) contains several images of the outer leaf of the nuclear membrane fusing with the inner surface of the intracytoplasmic membrane at pore-like spots. See also, Fuerst (1995: fig. 5). The intracytoplasmic membrane makes as sensible a homologue for the ER system as the purely hypothetical alternative possibilities. Another interesting ultrastructural point is the comparison between eukaryotic nuclear pores (see above) and the "crateriform structures" image from Brochier (2002). Notice, in particular, the structures towards the top of Brochier's image which appear to have been squeezed out of the membrane during preparation. Although the resemblance is impressive, it's hard to judge its significance. Once again, we caution that this is a different membrane plasma) from a different planctomycete (Pirellula).
Mans et al. (2004) have published a wonderful review of the nuclear pore complex and its possible origins. Their phylogenetic comparison fails to find any group of Eubacteria which has a particularly close relationship to the eukaryotes. Their analysis includes Planctomycetes. However, the significance and continued viability of this analysis is up in the air. Although the complete genome of Rhodopirellula was available at the time of the Mans review, the Kuenenia and Gemmata genomes were not. So, for example, Mans et al. state that planctomycetes lack the important pore structural element gle2. In fact, gle2 or a close homologue, is present in Gemmata. Fuerst (2005).
Similarly, NTF2, said to be absent from planctomycetes, turns up in Kuenenia with several very respectable matches on Superfamily. The gene has been annotated as coding for the structurally very similar) Δ-5-3-ketosteroid isomerase. This enzyme is known largely from unusual bacterial parasites who, like young lawyers, can survive solely on a diet of testosterone. This seems an unlikely sort of enzyme for Kuenenia, which is not widely known for its aggressive litigation tactics. Thus, we think the protein is probably an NTF2 homologue instead. Inspired by this information, we also took a small portion of the enormous mouse RanB2 sequence (a portion selected because it was reported to be similar to RanB1) from Wu et al. (1995) and searched against the incomplete data on Gemmata. This turned up a reasonable (33% identical, score = 55) match on a protein basis.
Of course none of this protein work has been done on a structural basis. Accordingly, we're a bit dubious about the whole thing. However, the point is not to nominate Gemmata as a candidate for the neomuran cenancestor. The point is that is that it has a mosaic of features and protein domains which suggest that it is one descendent of a radiation which included the Neomura, a radiation which was based close to, or conceivably inside, the crown group Planctomycetes -- except, of course, for those revolting proteasomes.
CONTINUED ON NEXT PAGE
ATW061129.
checked ATW061130, edited RFVS111206
 We have briefly discussed proteasomes elsewhere. This is the second of the rough spots to get over. As noted in the glossary entry at the link, proteasomes are related to chaperonins, such as GroEl. However, the function of the proteasome is to destroy proteins, rather than to rehabilitate them. For the Planctomycetes, this this is not only a rough spot. It may be the end of the road.
We have briefly discussed proteasomes elsewhere. This is the second of the rough spots to get over. As noted in the glossary entry at the link, proteasomes are related to chaperonins, such as GroEl. However, the function of the proteasome is to destroy proteins, rather than to rehabilitate them. For the Planctomycetes, this this is not only a rough spot. It may be the end of the road.
 This would be rather discouraging -- and impossible to reconcile fully with the rest of the evidence -- except that we may have overstated the case. While it is true that the 20S actinobacterial proteasomes are quite similar to their neomuran counterparts, the Gram-negative bacteria have HslV complexes which are almost as similar. True, they form rings which are hexameric, rather than heptameric; but they are not really so different for all that
This would be rather discouraging -- and impossible to reconcile fully with the rest of the evidence -- except that we may have overstated the case. While it is true that the 20S actinobacterial proteasomes are quite similar to their neomuran counterparts, the Gram-negative bacteria have HslV complexes which are almost as similar. True, they form rings which are hexameric, rather than heptameric; but they are not really so different for all that  Nothing could be more obvious about the Eukarya than their internal membrane system. The nucleus, endoplasmic reticulum and Golgi apparatus make the eukaryotic cell instantly recognizable. Likewise, the internal compartmentation of planctomycetes also makes them instantly recognizable among bacteria. However, this comparison is potentially misleading in at least two ways.
Nothing could be more obvious about the Eukarya than their internal membrane system. The nucleus, endoplasmic reticulum and Golgi apparatus make the eukaryotic cell instantly recognizable. Likewise, the internal compartmentation of planctomycetes also makes them instantly recognizable among bacteria. However, this comparison is potentially misleading in at least two ways. Although internal membrane systems are common enough in bacteria, internal membranes which wall off the chromosome are not. In fact, such membranes are unique to the Planctomycetes. That's not surprising, since it creates an awkward problem. Bacteria couple transcription with translation. That is, protein is synthesized while RNA is still bound to the chromosome. How, then, does one supply the cell wall, plasma membrane and external cytoplasm with protein? Evidently, planctomycetes have some sort of post-translational transport mechanism which can carry protein through the intracytoplasmic membrane and direct it to the appropriate destination.
Although internal membrane systems are common enough in bacteria, internal membranes which wall off the chromosome are not. In fact, such membranes are unique to the Planctomycetes. That's not surprising, since it creates an awkward problem. Bacteria couple transcription with translation. That is, protein is synthesized while RNA is still bound to the chromosome. How, then, does one supply the cell wall, plasma membrane and external cytoplasm with protein? Evidently, planctomycetes have some sort of post-translational transport mechanism which can carry protein through the intracytoplasmic membrane and direct it to the appropriate destination.
 In fact, cytochrome c peroxidase turns out to a strapping example of an α-solenoid system. Even the small β domains (in blue) take on the correct antiparallel, triangular appearance of "propeller" blades when viewed from an appropriate angle. Bear in mind, however, that this is the wrong planctomycete (Keunenia, rather than Gemmata) and the wrong membrane (anammoxosome, not nuclear).
In fact, cytochrome c peroxidase turns out to a strapping example of an α-solenoid system. Even the small β domains (in blue) take on the correct antiparallel, triangular appearance of "propeller" blades when viewed from an appropriate angle. Bear in mind, however, that this is the wrong planctomycete (Keunenia, rather than Gemmata) and the wrong membrane (anammoxosome, not nuclear). 