Origins of the Eukarya - 5
 We have relatively little to say here, since not a great deal is known about the internal membrane systems of bacteria (for a nice review of the eukaryote system, see Bonifacino & Glick, 2004). As discussed earlier, internal membrane structures are relatively commonplace in Eubacteria. What distinguishes these from a eukaryotic reticulum is the addition of a well-developed cytoskeleton. For the reasons discussed in the cytoskeleton section, baritones should therefore be, by far, the most promising candidates to develop this eukaryotic feature.
We have relatively little to say here, since not a great deal is known about the internal membrane systems of bacteria (for a nice review of the eukaryote system, see Bonifacino & Glick, 2004). As discussed earlier, internal membrane structures are relatively commonplace in Eubacteria. What distinguishes these from a eukaryotic reticulum is the addition of a well-developed cytoskeleton. For the reasons discussed in the cytoskeleton section, baritones should therefore be, by far, the most promising candidates to develop this eukaryotic feature.
In addition, Devos et al. (2004) have pointed out that the components of the nuclear pore complex and the proteins involved in the coated vesicle complex are very likely related. Thus, the remarkable development of the baritone cytoskeleton plus the available information on nuclear pore complex in planctomycetes suggest that we are looking in the right place for the origins of the endoplasmic reticulum as well. The one, essentially irrefutable, fact about Gemmata is that it must have an uncoupled translation system and must transport RNA through the nuclear membrane. Given those facts, something must be acting in a manner analogous to a nuclear pore + endoplasmic reticulum system as a matter of topological necessity. Perhaps the Gemmata system is entirely different, but it must have some system to perform these functions -- which is more than one can say for any other sort of bacterium.
 Cavalier-Smith gives pride of place to the Golgi complex, since this membrane complex is particularly important in trafficking with the outside world. This is critical to his approach, since he emphasizes the significance of mitochondrial "helotization." Cavalier-Smith starts with Actinobacteria, so he needs a way to acquire tenor/baritone genes early in the game. He does this by positing early acquisition of mitochondria, which requires phagocytosis of an α-proteobacterium, which in turn probably requires a Golgi apparatus. Thus, his Golgi apparatus evolves very early, with secondary loss of everything in the Archaea. In contrast, we'd speculate that the Golgi actually came last, particularly if Gemmata is the more appropriate model. This sequence of events is consistent with the evidence discussed above, the absence of anything Golgi-like in Archaea, and also with data from basal eukaryotes, a few of which do not have a Golgi complex at all. Simpson et al. (2002).
Cavalier-Smith gives pride of place to the Golgi complex, since this membrane complex is particularly important in trafficking with the outside world. This is critical to his approach, since he emphasizes the significance of mitochondrial "helotization." Cavalier-Smith starts with Actinobacteria, so he needs a way to acquire tenor/baritone genes early in the game. He does this by positing early acquisition of mitochondria, which requires phagocytosis of an α-proteobacterium, which in turn probably requires a Golgi apparatus. Thus, his Golgi apparatus evolves very early, with secondary loss of everything in the Archaea. In contrast, we'd speculate that the Golgi actually came last, particularly if Gemmata is the more appropriate model. This sequence of events is consistent with the evidence discussed above, the absence of anything Golgi-like in Archaea, and also with data from basal eukaryotes, a few of which do not have a Golgi complex at all. Simpson et al. (2002).
While we're in that part of phylospace, we might briefly consider the lowly nucleomorph. A few basal protists have acquired the benefits of photosynthesis by engulfing green algae. Some of these algal organelles retain a very basic nucleus and endoplasmic reticulum. As shown in the image from Protist Image Data, these have a peculiar gestalt resemblance to the Gemmata system with its unadorned nuclear membrane, simple pores, and unreticulated intracellular membrane. This isn't evidence of anything, of course -- but a thought-provoking image all the same.
 The neomuran system for translation from mRNA is "remarkably different" from the eubacterial system. Woese 2002). In fact, they are so different that it is rather difficult to tease a phylogenetic signal out of the data which will let us distinguish among Eubacteria. We know of only two exceptions. The first exception we have already discussed. Some planctomycetes have uncoupled mRNA translation from transcription. All of Gemmata's DNA is in the nuclear compartment. Considerable amounts of RNA are located outside the nuclear compartment. Lindsay et al. (2001). So, unless this RNA is just hanging around looking bored, the chances are that it is making protein.
The neomuran system for translation from mRNA is "remarkably different" from the eubacterial system. Woese 2002). In fact, they are so different that it is rather difficult to tease a phylogenetic signal out of the data which will let us distinguish among Eubacteria. We know of only two exceptions. The first exception we have already discussed. Some planctomycetes have uncoupled mRNA translation from transcription. All of Gemmata's DNA is in the nuclear compartment. Considerable amounts of RNA are located outside the nuclear compartment. Lindsay et al. (2001). So, unless this RNA is just hanging around looking bored, the chances are that it is making protein.
A more subtle signal can be detected from the peculiar amino acid hypusine. Cavalier-Smith (2002a) aptly describes its role as follows:
Neomura have a novel set of elongation factors (eIF-2) in addition to the universal IF-2 factors; both kinds are involved in forming the complex of the charged initiator tRNA with the mRNA and small ribosomal subunit. Neomura alone have an eIF-2A responsible for dissociating the two ribosomes, an eIF-2B responsible for GTP recycling on eIF2 and an eIF-5A. The latter is particularly significant as the only protein in the living world with the amino acid hypusine. Since hypusine is modified from lysine by two successive enzymic steps, effected by proteins, this is compelling evidence that hypusine and the neomuran eIF-5A that depends on it are derived neomuran characters and that the simpler eubacterial system is ancestral.
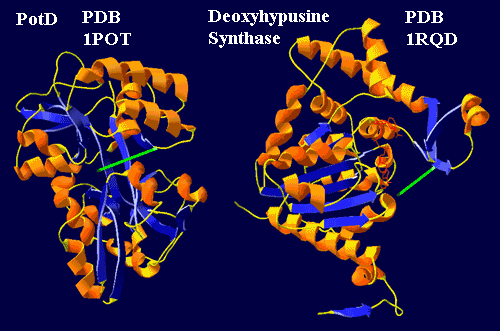 One of the two enzymes responsible for making hypusine is deoxyhypusine synthase. Oddly enough, a gene for this enzyme, or something very like it, is found in a handful of bacteria: they are all Gram negative tenors, and basses, and the baritones Rhodopirellula and Gemmata. Brochier et al. (2004) [18]. Brochier et al. find that the planctomycete sequences are particularly close to those of basal Archaea. This fact they take as unmistakable evidence of HGT, even though, in the very same paragraph, they admit that the eubacterial sequences are monophyletic...
One of the two enzymes responsible for making hypusine is deoxyhypusine synthase. Oddly enough, a gene for this enzyme, or something very like it, is found in a handful of bacteria: they are all Gram negative tenors, and basses, and the baritones Rhodopirellula and Gemmata. Brochier et al. (2004) [18]. Brochier et al. find that the planctomycete sequences are particularly close to those of basal Archaea. This fact they take as unmistakable evidence of HGT, even though, in the very same paragraph, they admit that the eubacterial sequences are monophyletic...
Brochier et al. express some uncertainty as to why a bacterium would have such an enzyme, since bacteria lack EF-5a. Perhaps we've missed something, but this seems to be the least mysterious thing about this enzyme. Any enzyme capable of transferring an aminobutyl group to a bound ω-aminoalkyl group (as in any bound lysine, spermine, putrescine, etc.) is sure of finding a rewarding career managing the polyamines in bacterial chromatin.
The story (admittedly a scenario) is not difficult to reconstruct. The main protein for trafficking in polyamines seems to be PotD. PotD works like a Venus flytrap, binding the substrate (usually spermidine, approximated by a green line), across a network of beta sheets. The end to the left in the figure is hydrogen bonded between two loops extending from the upper and lower "jaws." Sugiyama et al. (1996). Deoxyhypusine synthase is likely to be a relative which once did the same job (Lee et al., 2001) and, in some bacteria, still probably performs a transport function. However, in the neomuran synthase, the beta structures have been reoriented to form a standard Rossman fold, which binds NAD. Consequently the "left" end of the spermidine is now very loosely bound. However, it is forced into contact with the nucleotide when the enzyme binds eIF-5A. That explains the extraordinary specificity of the enzyme. That is, the synthase may bind any number of proteins, but only one is exactly the right shape to force the wayward spermidine into appropriate contact with NAD. That contact shorts out the high energy electron in NAD, cuts the spermidine in two, and welds one half onto the protein substrate -- an explanation extrapolated and grossly oversimplified from Umland et al. (2004). Thus, in a reasonably understandable way, transport evolved into transformation.
The hypusine story is actually part of a much bigger biochemical trend. Neomura often create novel functional groups by post-translational modification of amino acids. This is relatively rare in Eubacteria. Cavalier-Smith (2002). Cavalier-Smith illustrates this point with a discussion of amino acid phosphokinases. These are particularly important in transcriptional regulation. As it happens, baritones and tenors are particularly well-supplied with these enzymes. Fuerst (2005); Jelsbak et al. (2005).
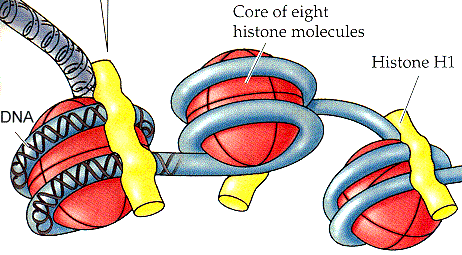 We normally think of eukaryotes as having condensed, histone-covered chromatin, while bacteria have fuzzy, "coralline" chromatin without histones. As mentioned earlier, there are a large number of exceptions. Bendich & Drlica (2000). Nevertheless, this isn't far from the case. Cavalier-Smith (2002) correctly predicted that all Neomuran lineages originally came with histones. Čuboňová et al. (2005). By contrast, Eubacteria have polyamines and HU proteins. The HU proteins are probable relatives of histone H1. Cavalier-Smith has often stated that the Actinobacteria have the most histone-like HU known (Cavalier-Smith, 2002). This may have been correct at one time, but is now very doubtful. The bass/baritone Chlamydiales probably own that title at this point. Griffiths et al. (2006).
We normally think of eukaryotes as having condensed, histone-covered chromatin, while bacteria have fuzzy, "coralline" chromatin without histones. As mentioned earlier, there are a large number of exceptions. Bendich & Drlica (2000). Nevertheless, this isn't far from the case. Cavalier-Smith (2002) correctly predicted that all Neomuran lineages originally came with histones. Čuboňová et al. (2005). By contrast, Eubacteria have polyamines and HU proteins. The HU proteins are probable relatives of histone H1. Cavalier-Smith has often stated that the Actinobacteria have the most histone-like HU known (Cavalier-Smith, 2002). This may have been correct at one time, but is now very doubtful. The bass/baritone Chlamydiales probably own that title at this point. Griffiths et al. (2006).
In any case, we decline to get involved in competing sequence trees. There are too many H1-like sequences, and far too many relatives of histone proteases, histone kinases, histone acetyltransferases, and other histone what-nots. By contrast, there are no reasonable bacterial relatives of the core histones themselves (i.e., all of the other histones). Without core histones, the presence or absence of H1 is essentially irrelevant. It is the core histones which form the characteristic histone nucleosome. The core histones bind DNA compactly in a supercoiled form. H1 simply continues to do what HU always did -- loosely stapling DNA regions together.
 Why does this matter to the cell? The usual explanations involve all sorts of complex stories about gene regulation and transcription. This is because such explanations are written by deep thinkers and competent experimentalists, like Cavalier-Smith, rather than failed lab monkeys, like us. The former group tend to design and execute successful experiments. Since their own experiments work, they naturally gravitate toward functional explanations on the unstated assumption that nature must also work in the same competent manner as do they. We know better. Based on our own, less happy, experiences, we tend to think in terms of basic chemistry, rather than function, and of the large number of ways in which simple things can go horribly wrong without ever functioning at all. Evolution doesn't generally favor the organism with the most sophisticated RNA polymerase co-factors. It favors the organism which survives being left out on the lab bench all night, unnoticed behind a coffee mug.
Why does this matter to the cell? The usual explanations involve all sorts of complex stories about gene regulation and transcription. This is because such explanations are written by deep thinkers and competent experimentalists, like Cavalier-Smith, rather than failed lab monkeys, like us. The former group tend to design and execute successful experiments. Since their own experiments work, they naturally gravitate toward functional explanations on the unstated assumption that nature must also work in the same competent manner as do they. We know better. Based on our own, less happy, experiences, we tend to think in terms of basic chemistry, rather than function, and of the large number of ways in which simple things can go horribly wrong without ever functioning at all. Evolution doesn't generally favor the organism with the most sophisticated RNA polymerase co-factors. It favors the organism which survives being left out on the lab bench all night, unnoticed behind a coffee mug.
So applying this Principle of Incompetence, let us consider some very basic chemistry. DNA isn't simply an information storage device. Before we can worry about the optimum functionality of the cell's data processing system, we have to deal with the fact that DNA is, first and foremost, a really big, charged polymer. If nothing else, the presence of so many exposed phosphate groups in dispersed DNA puts a significant limit on the size of the genome a bacterium can support. There is nothing subtle about this. Long charged polymers of any kind bind water -- and anything else polar -- into a viscous glop with high osmotic pressure. Worse, DNA glop (as we know from sad personal experience) is also highly vulnerable to damage by shearing, nucleases, radiation, and being looked at funny.
Both planctomycetes and various Actinobacteria are notable for being rather small cells with some of the largest genomes in the Eubacteria. By cramming almost 107 base pairs of DNA into cells about 1 micron in diameter, planctomycetes are, very literally, pushing the envelope on DNA content. Certainly, the planctomycetes in particular need as much DNA as they can get because they are oligotrophs. They make their living surviving the natural equivalent of being dropped on the floor, contaminated with yeast, and being left on the lab bench all night. Precisely because things can go horribly wrong in so many unpredictable ways, bugs who live on scraps in marginal environments need extensive metabolic flexibility. Yet the DNA concentration required for this flexibility is over 2% w/v -- somewhat higher than the concentration of DNA in a typical eukaryotic cell, and flatly impossible without special packaging. We don't have to speculate whether planctomycete chromatin is supercoiled, condensed, and packaged into orderly, charge-shielded arrays. Nothing else could prevent these cells, and a sizeable volume of the surrounding medium, from being converted into solid chunks of hydrated glop.
CONTINUED ON NEXT PAGE
ATW061129.
checked ATW061201, edited RFVS111204
 We have relatively little to say here, since not a great deal is known about the internal membrane systems of bacteria (for a nice review of the eukaryote system, see Bonifacino & Glick, 2004). As discussed earlier, internal membrane structures are relatively commonplace in Eubacteria. What distinguishes these from a eukaryotic reticulum is the addition of a well-developed cytoskeleton. For the reasons discussed in the cytoskeleton section, baritones should therefore be, by far, the most promising candidates to develop this eukaryotic feature.
We have relatively little to say here, since not a great deal is known about the internal membrane systems of bacteria (for a nice review of the eukaryote system, see Bonifacino & Glick, 2004). As discussed earlier, internal membrane structures are relatively commonplace in Eubacteria. What distinguishes these from a eukaryotic reticulum is the addition of a well-developed cytoskeleton. For the reasons discussed in the cytoskeleton section, baritones should therefore be, by far, the most promising candidates to develop this eukaryotic feature.

 One of the two enzymes responsible for making hypusine is deoxyhypusine synthase. Oddly enough, a gene for this enzyme, or something very like it, is found in a handful of bacteria: they are all Gram negative tenors, and basses, and the baritones Rhodopirellula and Gemmata.
One of the two enzymes responsible for making hypusine is deoxyhypusine synthase. Oddly enough, a gene for this enzyme, or something very like it, is found in a handful of bacteria: they are all Gram negative tenors, and basses, and the baritones Rhodopirellula and Gemmata.  We normally think of eukaryotes as having condensed, histone-covered chromatin, while bacteria have fuzzy, "coralline" chromatin without histones. As mentioned
We normally think of eukaryotes as having condensed, histone-covered chromatin, while bacteria have fuzzy, "coralline" chromatin without histones. As mentioned  Why does this matter to the cell? The usual explanations involve all sorts of complex stories about gene regulation and transcription. This is because such explanations are written by deep thinkers and competent experimentalists, like Cavalier-Smith, rather than failed lab monkeys, like us. The former group tend to design and execute successful experiments. Since their own experiments work, they naturally gravitate toward functional explanations on the unstated assumption that nature must also work in the same competent manner as do they. We know better. Based on our own, less happy, experiences, we tend to think in terms of basic chemistry, rather than function, and of the large number of ways in which simple things can go horribly wrong without ever functioning at all. Evolution doesn't generally favor the organism with the most sophisticated RNA polymerase co-factors. It favors the organism which survives being left out on the lab bench all night, unnoticed behind a coffee mug.
Why does this matter to the cell? The usual explanations involve all sorts of complex stories about gene regulation and transcription. This is because such explanations are written by deep thinkers and competent experimentalists, like Cavalier-Smith, rather than failed lab monkeys, like us. The former group tend to design and execute successful experiments. Since their own experiments work, they naturally gravitate toward functional explanations on the unstated assumption that nature must also work in the same competent manner as do they. We know better. Based on our own, less happy, experiences, we tend to think in terms of basic chemistry, rather than function, and of the large number of ways in which simple things can go horribly wrong without ever functioning at all. Evolution doesn't generally favor the organism with the most sophisticated RNA polymerase co-factors. It favors the organism which survives being left out on the lab bench all night, unnoticed behind a coffee mug.