Origins of the Eukarya - 2
 What the Eukarya do have are outer membrane proteins, particularly porins. We pause here, stoically, to endure the usual whining about lack of sequence homology or HGT. ... If you are quite finished now, we will move on. Eukaryotic porins are strikingly similar the porins of bacterial tenors and baritones. It turns out that sequence is about the only feature they do not share. They both perform the same class of functions, i.e., making pores so that molecules, particularly charged molecules, can enter or leave the cell through the apolar membrane. They are all based on a very similar β-barrel structure and sometimes share strikingly similar structural details and molecular mechanisms. See, e.g., Bishop et al. (2005); Reumann et al. 1998); Iyer & Delcour (1997). But the clincher, to our way of thinking, is that bacteria can actually substitute their own porins for those of a eukaryotic host during infection, using the host cell's own porin-inserting program. Müller et al. (2002).
What the Eukarya do have are outer membrane proteins, particularly porins. We pause here, stoically, to endure the usual whining about lack of sequence homology or HGT. ... If you are quite finished now, we will move on. Eukaryotic porins are strikingly similar the porins of bacterial tenors and baritones. It turns out that sequence is about the only feature they do not share. They both perform the same class of functions, i.e., making pores so that molecules, particularly charged molecules, can enter or leave the cell through the apolar membrane. They are all based on a very similar β-barrel structure and sometimes share strikingly similar structural details and molecular mechanisms. See, e.g., Bishop et al. (2005); Reumann et al. 1998); Iyer & Delcour (1997). But the clincher, to our way of thinking, is that bacteria can actually substitute their own porins for those of a eukaryotic host during infection, using the host cell's own porin-inserting program. Müller et al. (2002).
You may object that the β-barrel porins at issue are located in mitochondrial membranes, and might have been imported with the proteobacterial DNA of the original mitochondrial symbiote. This is made less likely precisely because mitochondrial porins have less sequence homology to bacterial porins than other eukaryotic proteins. In fact, by sequence, they are entirely eukaryotic. In any case, the distribution of these porins -- know in the biomed trade as "VDACs" (voltage-dependent anion channels) -- is not restricted to the mitochondrial outer membrane. Sun & Liao (2002)[3]; Buettner et al. (2000); see, generally, Garrow et al. (2005).
 Here's the point of all this. The complete genome sequence reveals that, while the baritone Rhodopirellula is not exactly a porin star, it has a fair number of these proteins. Outer Membrane Channels Site. This is true despite the absence of a conventional outer membrane. But what about the altos? They have no porins at all. Or, to be completely accurate, they have no porins which have any kind of homology to those in any other taxon, bacterial or otherwise, as Cavalier-Smith (2006) admits.
Here's the point of all this. The complete genome sequence reveals that, while the baritone Rhodopirellula is not exactly a porin star, it has a fair number of these proteins. Outer Membrane Channels Site. This is true despite the absence of a conventional outer membrane. But what about the altos? They have no porins at all. Or, to be completely accurate, they have no porins which have any kind of homology to those in any other taxon, bacterial or otherwise, as Cavalier-Smith (2006) admits.
Another important outer membrane biochemical is Lipid A. We won't bother with structural details, save to note that Lipid A is a rather distinctive and is unique to Gram negative bacteria -- almost. Armstrong et al. (2002) have recently reported Lipid A in Chlorella, a green alga. Green plants, to judge from Arabidopsis, apparently have the full complement of enzymes needed to synthesize Lipid A. Wu et al. 2004). These "bacterial" genes are in the nucleus, and the lipid is located in the cell membrane, so that the gene products are also presumably located in the cell's own membranes. Once again, it is entirely possible that these genes could have been picked up from an organelle -- in this case the chloroplast. However, they cannot have been derived from an actinobacterial alto, because altos have no such genes. Planctomycetes, as you may have guessed, have most or all of these genes and may even produce some Lipid A [4]. Fuerst (2005); Jenkins et al. (2002). See also Krupa & Srinivasan (2002).
 At this point, you have probably picked up the pattern. If Eukarya share some feature with altos, Cavalier-Smith tends to ascribe it to vertical inheritance; but if the feature is uniquely shared with baritones, he invokes horizontal transfer from organelles. However, it is harder to run this game with absence characters. Actinobacterial altos have a particularly thick murein cell wall. Planctomycete baritones share with eukaryotes the absence of a peptidoglycan cell wall. In fact, penicillin, which blocks cell wall synthesis, can be used to isolate planctomycetes. Rhodopirellula's genome contains some of the genes for making peptidoglycans, but not a full set. Glöckner et al. (2003). Like Archaea and Eukarya, Planctomycetes make heavy use of glycoproteins
[5]instead.
At this point, you have probably picked up the pattern. If Eukarya share some feature with altos, Cavalier-Smith tends to ascribe it to vertical inheritance; but if the feature is uniquely shared with baritones, he invokes horizontal transfer from organelles. However, it is harder to run this game with absence characters. Actinobacterial altos have a particularly thick murein cell wall. Planctomycete baritones share with eukaryotes the absence of a peptidoglycan cell wall. In fact, penicillin, which blocks cell wall synthesis, can be used to isolate planctomycetes. Rhodopirellula's genome contains some of the genes for making peptidoglycans, but not a full set. Glöckner et al. (2003). Like Archaea and Eukarya, Planctomycetes make heavy use of glycoproteins
[5]instead.
There have been some very recent reports (i.e. we haven't actually read the papers yet) claiming peptidoglycan synthesis in eukaryotic cells. It will be interesting to see if this product is truly murein, or some partial product, similar to what one might expect from wall-less baritone bacteria. We should emphasize that N-acetyl-glucosamine, one of the key ingredients in murein, remains an important part of planctomycete metabolism, as in eukaryotes. Indeed, planctomycetes can survive on N-acetyl-glucosamine as their sole source of carbon and nitrogen. Jenkins et al. (2002).
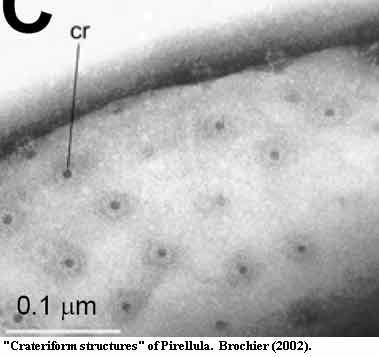
The planctomycete cell walls have a couple of additional weird features we'll discuss a bit later. These include some rounded protrusions (sometimes) and "crateriform structures" (always). The latter are shown particularly well in the figure from Brochier (2002). No one knows what these are at the moment. Look carefully at the construction of the toroidal rims of the "craters" and perhaps ruminate a bit...
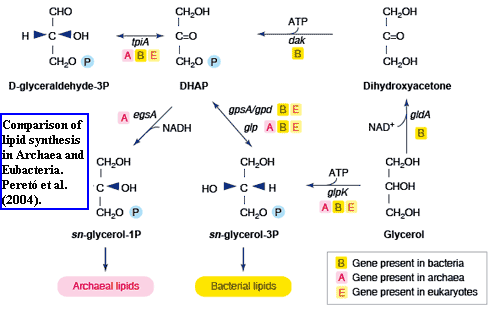 We have now reached one of two rough spots on the road. For the most part, the baritones make remarkably good proto-sopranos. However, they may fail in two important areas: lipid stereochemistry [7] and proteasomes. In brief, the lipid stereochemistry story is shown in the figure from Peretó et al. (2004). It goes like this.
We have now reached one of two rough spots on the road. For the most part, the baritones make remarkably good proto-sopranos. However, they may fail in two important areas: lipid stereochemistry [7] and proteasomes. In brief, the lipid stereochemistry story is shown in the figure from Peretó et al. (2004). It goes like this.
The backbone of bulk cell membrane lipids is always glycerol, a three-carbon sugar. Glycerol, in the form of glycerol phosphate, is made from dihydroxyacetone phosphate ("DHAP" in the diagram). Notice that the middle carbon in glycerol phosphate is attached to four different things. This means that it is asymmetrical and has mirror-image forms which are not equivalent (enantiomers): glycerol-1-phosphate ("G1P") and glycerol-3-phosphate ("G3P"). Archaea create the G1P form, using G1P dehydrogenase (G1PDH), while everyone else creates G3P, using G3P dehydrogenase (G3PDH). The sticky part is that, the two enzymes are very different by sequence. The Actinobacterial altos have a perfectly good G1PDH, like the Archaea, in addition to their G3PDH. We are told that the tenors and baritones lack this enzyme. Forterre (2006); Peretó et al. (2004). Cavalier-Smith 2006) makes much of this -- and well he should. It's a good point.
It is not, however, quite good enough, for two reasons. The first, and less important, point is that there are excellent reasons to think that sequence analysis overstates the difference between G1PDH and G3PDH. I include this item only as an example of how sequence phylogenies can fail. I do not argue that Baritones could somehow transmute
 one into the other. The second, and much more significant, point is that G1PDH is extremely similar to
3-dehydroquinate synthase (DHQS), an enzyme with which baritones are generously endowed.
one into the other. The second, and much more significant, point is that G1PDH is extremely similar to
3-dehydroquinate synthase (DHQS), an enzyme with which baritones are generously endowed.
G1PDH and G3PDH are both members of the incredibly prolific dehydrogenase family of enzymes. Both enzymes have two distinct domains separated by a deep catalytic cleft. Both include a dehydrogenase domain, coupled to a nucleotide-binding domain with a distinctive series of alternating, extended, α-helix-β-sheet sequences known as a Rossman fold. The nucleotide-binding binds, variously, NADH, NADPH, or FADH, common nucleotide electron transport carriers which donate the proton needed to reduce the substrate [6]. The substrate may be, as in our case, DHAP. In some related enzymes it may be any of several intermediates in the metabolism of sterols, hopanoids, aromatic amino acids, or glycoproteins. All dehydrogenases appear to be phylogenetically related, and the lineage probably extends back before LUCA. See Peretó et al. (2004) for a review.
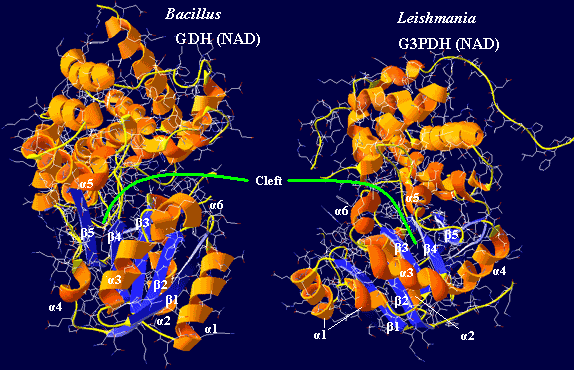
No one has determined the crystal structure of G1PDH. However, Dr. Yosuke Koga of the University of Occupational and Environmental Health in Kitakyushu, Dr. Jin-Suk Han of the Dongeui Institute of Technology in Busan, and others have tried to develop the structure by modeling. Han & Ishikawa (2005); Koga et al. (2003); Daiyasu et al. (2002). To make a long story short, these workers deduced (and have partially confirmed) that the structure of G1PDH ought to be almost the same as that of glycerol dehydrogenase (GDH, yet another member of the dehydrogenase class), but G1PDH is arranged so as to extract a proton from the opposite side of the nucleotide as compared to G3PDH.[8]
 Armed with this data, we betook ourselves to the PDB site and used DeepView to generate comparable images of G3PDH from a eukaryote, Leishmania (PDB
1EVZ), and GDH from a bacterium, Bacillus (PDB
1JQA). The results are shown in the figure. The substrate binding domains (top) really are rather different. The really striking part is the incredible similarity of the nucleotide binding domains, as the papers cited above noted. We have arbitrarily labeled the homologous elements of the two. Despite the enormous phylogenetic gap between the Leishmania and Bacillus, the nucleotide-binding domains are very similar, except that they are mirror images! Thus we have "enantiomeric" nucleotide binding domains for enantiomeric substrates.
Armed with this data, we betook ourselves to the PDB site and used DeepView to generate comparable images of G3PDH from a eukaryote, Leishmania (PDB
1EVZ), and GDH from a bacterium, Bacillus (PDB
1JQA). The results are shown in the figure. The substrate binding domains (top) really are rather different. The really striking part is the incredible similarity of the nucleotide binding domains, as the papers cited above noted. We have arbitrarily labeled the homologous elements of the two. Despite the enormous phylogenetic gap between the Leishmania and Bacillus, the nucleotide-binding domains are very similar, except that they are mirror images! Thus we have "enantiomeric" nucleotide binding domains for enantiomeric substrates.
Here's the lesson. The nucleotide-binding domains of G1PDH and G3PDH, however similar, will have little sequence homology, since the sequence of amino acids of one will be essentially the reverse of the other. Thus, it isn't particularly surprising, and is essentially meaningless, that the two proteins are "unrelated" by sequence. It's the structure that really counts, not the sequence. Sequence can often be used as a rough proxy for structure; but in other cases, such as this one, it can be misleading.
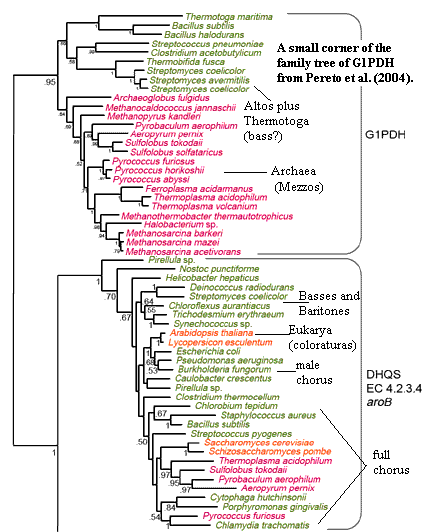
All that doesn't get us anywhere, since it doesn't explain how baritones might have developed a G1PDH which is not present in their living representatives. However, Peretó's sequence analysis shows that G1PDH is much more closely related to another enzyme, 3-dehydroquinate synthase (DHQS), a ubiquitous protein close to the base of half a dozen critical metabolic pathways. In fact, DHQS was the enzyme most closely related to G1PDH by sequence, and Pirellula sp. strain 1 (now Rhodopirellula) had the DHQS which was most closely related to G1PDH of all of the DHQS sequences tested.
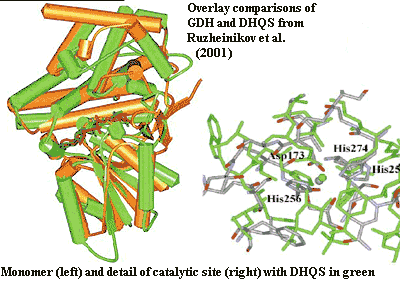
However, the sequence similarity between G1PDH and DHQS grossly understates the case. Several years ago, two independent groups of workers, using entirely different techniques, had already realized that GDH and DHQS were "remarkably similar" by structure and catalytic mechanism [10]. Bartlett et al. (2003); Ruzheinikov et al. (2001). In fact, as both groups observed, a substantial portion of the catalytic site is virtually identical. Nor is this a coincidence. DHQS is a multi-step enzyme. The first step is the NADH and Zn++-instigated attack of a water molecule to oxidize a hydroxyl to a ketone. DHQS then goes on to do other things. That first step is precisely what GDH does, except that it accepts a smaller substrate and stops after that step. If an innocent little glycerol molecule accidentally wandered into the active site of a negligent DHQS (and it might, because DHQS accepts a much larger, complex 7-carbon sugar as substrate), it would emerge as dihydroxyacetone, just as in GDH. The DHQS could not perform its additional functions and the DHA would be released in that case, because glycerol doesn't have the other structures on which the remaining steps of the DHQS reaction operate.
Given the conclusion of Koga, Han, and co-workers that G1PDH is a structural clone of GDH, it then seems that G1PDH, GDH and DHQS form a very closely related group. Given two billion years to play with, it simply can't be that unlikely that a DHQS was exapted to act as a G1PDH. The "patchy" distribution of G1PDH among the Eubacteria -- Actinobacteria altos), some Proteobacteria (tenors), and
 Thermotoga who knows?) -- is not evidence of HGT, contra Martin & Russell (2002); Peretó et al. (2004). It is not evidence that Actinobacteria are the sisters of Neomura, contra Cavalier-Smith (2006). It is probably just evidence that a workable G1PDH is simply not that hard to cobble together from relatively common spare parts.
Thermotoga who knows?) -- is not evidence of HGT, contra Martin & Russell (2002); Peretó et al. (2004). It is not evidence that Actinobacteria are the sisters of Neomura, contra Cavalier-Smith (2006). It is probably just evidence that a workable G1PDH is simply not that hard to cobble together from relatively common spare parts.
That leads us to the next subject: sterols. Most bacteria do not make sterols. They make hopanoids, which are chemically similar and may do roughly the same primary job, controlling membrane viscosity.Several groups have published reports of sterol biosynthesis in various bacteria. Most have turned out to be wrong. Brocks et al. (2003). Cavalier-Smith (2006) no longer claims sterol synthesis as strong indicator of the Actinobacterial origin of eukaryotes, since sterols are plainly present in some tenors, particularly the methylotrophs. Planctomycetes also produce sterols. The interesting feature here is that the synthetic pathway in Gemmata is the shortest known sterol pathway in all of life. Fuerst (2005); Pearson et al. (2003). Gemmata is also probably the only anaerobe of any kind to make sterols. Primitive characters are normally not the best indicators of phylogeny, but, when the "primitive state" is otherwise unknown, its presence is suggestive. At the least, it helps exclude the possibility of HGT.
Know that we are prepared to drone on about the plasma membrane almost indefinitely. In fact, it might serve Cavalier-Smith right if someone were to write papers even more vast than his, commenting on his work. Then he might have to exhaust his own patience and overload his own optic tectum in the same way that others must when reading his offerings. Sadly, it seems unlikely that he will see this page; and it would be churlish for us to exact such a heavy price from you, just on the off-chance that you might turn out to be one particularly expansive Oxford biologist. Thus we will have to be 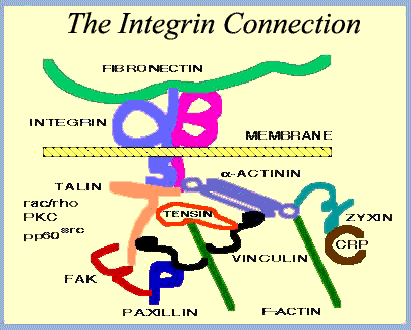 satisfied with a few more bullet points.
satisfied with a few more bullet points.
- Rhodopirellula, like neomurans, has an unreasonable number (1271) of sequences apparently coding for signal peptides, as well as a large array of secretion and cell surface proteins. Fuerst (2005); Glöckner et al. (2003).
- In aerobic Planctomycetes, the primary phospholipid components are palmitic, palmitoleic and oleic acids, "a pattern more typical of microeukaryotes than of Eubacteria." Fuerst (1995).
- Some (but not all) Planctomycetes and Verrucomicrobia (other baritones) appear to have integrins. In eukaryotes, integrins bind to microfilaments and the extracellular matrix and are important agents of signal transduction and motility. Fuerst (2004); Glöckner et al. (2003), Jenkins et al. (2002a). This is particularly interesting, as a) integrins are held together by disulfide bridges (Chillarón et al., 2001), and baritones are known for having many of these on the cell surface (Glöckner et al., 2003); plus (b) the motility bit is usually carried out in association with actin (Rose, 2006), which we'll take up later.
- Other "eukaryotic" actin-associated cell adhesion/motility proteins, cadherin and laminin G, are represented by several domains in Rhodopirellula. Zelensky & Gready (2005).
- Cavalier-Smith (2002, 2002a, 2006) repeatedly states that Actinobacteria (not altos in general) uniquely share phosphatidyl inositol lipid with the Eukarya (Archaea have the analogous myo-inositol). In fact, tenors and baritones also have enzymes specific to phosphatidyl inositol regulation -- not quite the same thing, but close. Pagliarini et al. (2004). See also IPR000403.
G-proteins are also critical signal transduction elements in eukaryotes. They are known from δ-Proteobacteria (tenors), but not altos. Cavalier-Smith (2002). Planctomycetes have an unusually large number of these elements. Fuerst (2005).
And now, having run out of bullets for the moment, we are forced to reload and move on to other things.
CONTINUED ON NEXT PAGE
ATW061129. checked ATW061130, edited RFVS111204
 What the Eukarya do have are outer membrane proteins, particularly porins. We pause here, stoically, to endure the usual whining about lack of sequence homology or HGT. ... If you are quite finished now, we will move on. Eukaryotic porins are strikingly similar the porins of bacterial tenors and baritones. It turns out that sequence is about the only feature they do not share. They both perform the same class of functions, i.e., making pores so that molecules, particularly charged molecules, can enter or leave the cell through the apolar membrane. They are all based on a very similar β-barrel structure and sometimes share strikingly similar structural details and molecular mechanisms. See, e.g., Bishop et al. (2005); Reumann et al. 1998); Iyer & Delcour (1997). But the clincher, to our way of thinking, is that bacteria can actually substitute their own porins for those of a eukaryotic host during infection, using the host cell's own porin-inserting program. Müller et al. (2002).
What the Eukarya do have are outer membrane proteins, particularly porins. We pause here, stoically, to endure the usual whining about lack of sequence homology or HGT. ... If you are quite finished now, we will move on. Eukaryotic porins are strikingly similar the porins of bacterial tenors and baritones. It turns out that sequence is about the only feature they do not share. They both perform the same class of functions, i.e., making pores so that molecules, particularly charged molecules, can enter or leave the cell through the apolar membrane. They are all based on a very similar β-barrel structure and sometimes share strikingly similar structural details and molecular mechanisms. See, e.g., Bishop et al. (2005); Reumann et al. 1998); Iyer & Delcour (1997). But the clincher, to our way of thinking, is that bacteria can actually substitute their own porins for those of a eukaryotic host during infection, using the host cell's own porin-inserting program. Müller et al. (2002).
 At this point, you have probably picked up the pattern. If Eukarya share some feature with altos, Cavalier-Smith tends to ascribe it to vertical inheritance; but if the feature is uniquely shared with baritones, he invokes horizontal transfer from organelles. However, it is harder to run this game with absence characters. Actinobacterial altos have a particularly thick murein cell wall. Planctomycete baritones share with eukaryotes the absence of a peptidoglycan cell wall. In fact, penicillin, which blocks cell wall synthesis, can be used to isolate planctomycetes. Rhodopirellula's genome contains some of the genes for making peptidoglycans, but not a full set.
At this point, you have probably picked up the pattern. If Eukarya share some feature with altos, Cavalier-Smith tends to ascribe it to vertical inheritance; but if the feature is uniquely shared with baritones, he invokes horizontal transfer from organelles. However, it is harder to run this game with absence characters. Actinobacterial altos have a particularly thick murein cell wall. Planctomycete baritones share with eukaryotes the absence of a peptidoglycan cell wall. In fact, penicillin, which blocks cell wall synthesis, can be used to isolate planctomycetes. Rhodopirellula's genome contains some of the genes for making peptidoglycans, but not a full set.  We have now reached one of two rough spots on the road. For the most part, the baritones make remarkably good proto-sopranos. However, they may fail in two important areas: lipid
We have now reached one of two rough spots on the road. For the most part, the baritones make remarkably good proto-sopranos. However, they may fail in two important areas: lipid  one into the other. The second, and much more significant, point is that G1PDH is extremely similar to
3-dehydroquinate synthase (DHQS), an enzyme with which baritones are generously endowed.
one into the other. The second, and much more significant, point is that G1PDH is extremely similar to
3-dehydroquinate synthase (DHQS), an enzyme with which baritones are generously endowed. Armed with this data, we betook ourselves to the
Armed with this data, we betook ourselves to the  Thermotoga who knows?) -- is not evidence of HGT, contra
Thermotoga who knows?) -- is not evidence of HGT, contra 
