Origins of the Eukarya - 3
 This part is much easier. You won't have to memorize a bunch of incomprehensible enzyme nicknames or vainly attempt to recall metabolic pathways you tried to memorize about six hours before the final -- and never thought about again. (Did you think we didn't know about that?) Even better, just about everyone agrees on the homologies: actin = MreB, or possibly ParM [13], and tubulin = FtsZ. We'll mention some other homologies, but those are the only two which count.
This part is much easier. You won't have to memorize a bunch of incomprehensible enzyme nicknames or vainly attempt to recall metabolic pathways you tried to memorize about six hours before the final -- and never thought about again. (Did you think we didn't know about that?) Even better, just about everyone agrees on the homologies: actin = MreB, or possibly ParM [13], and tubulin = FtsZ. We'll mention some other homologies, but those are the only two which count.
For that matter even these homologies may not count all that much. The Archaea don't have actin or tubulin. Like other bacteria, they get by on MreB and FtsZ. Possibly, actin and tubulin were lost in the Archaea. That sounds unlikely, until you remember that early Archaea, like modern Archaea, were probably extremophiles. Lots of otherwise very useful proteins simply don't work well at, for example, pH 2 and 90° C. When attempting to swim in boiling acid, one tends to focus on essentials, forgoing even cell phones.
Bacterial division almost always depends on FtsZ, together with other proteins of the Fts group. We don't need to know much about them. In brief, FtsZ forms a circular belt around the cell, the Z-ring. During cell division, the belt is cinched tighter and tighter -- until the cell splits in two. FtsZ also has something to do with initiating DNA replication and seems to signal other necessary steps in cell division. What matters to us is the Z-ring belt. Like tubulin, FtsZ forms long, mechanically powerful, GTP-dependent polymers.
Molecular bean-counters tend to grumble about lack of sequence identity, but FtsZ and tubulin are almost identical by structure. That's significant because the structure is a bit out of the common run. The GTP-binding domain is a fairly ordinary Rossman fold. It is said to be slightly funny-looking by true aficionados of Rossman folds, but we need not rest on such effete discriminations. The Rossman fold is separated from the structural domain by an extremely long alpha helix (the "core helix"), which runs roughly parallel to the fan of β-sheets in the Rossman fold. The structural domain consists of four parallel β-sheets (one is actually anti-parallel) sticking out sideways from the core helix, with a couple of shorter helices wrapped loosely around the far end. The Rossman fold also bears a variably long tail with a helix at the end, like a nanoscale Ankylosaurus. Löwe & Amos (1998); van den Ent et al. (2001).
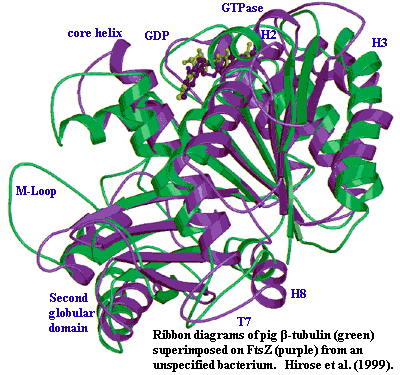
 FtsZ is, obviously, not functionally identical to tubulin. It has an entirely different sequence from tubulin. It does not use its GTPase in quite the same way; and it forms a associates with other strands in a different way. However, it is practically a duplicate of tubulin by structure, as the figure indicates. Hirose et al. (1999).
FtsZ is, obviously, not functionally identical to tubulin. It has an entirely different sequence from tubulin. It does not use its GTPase in quite the same way; and it forms a associates with other strands in a different way. However, it is practically a duplicate of tubulin by structure, as the figure indicates. Hirose et al. (1999).
Accordingly we know, in a general sense, where tubulin came from: FtsZ -- but whose FtsZ? Oddly enough, the answer lies in an absence character. FtsZ is everywhere. Over the entire span of life there are only five groups of organisms which lack FtsZ. Two are the eukaryotes (excluding plastid FtsZ) and one of the main groups of Archaea, the Crenarchaeota. All three others are baritones, including at least one of the planctomycetes. Marrington et al. (2004). How does that help? Let us add another fact. Only one eubacterium is known to have an actual tubulin. This is Prosthecobacter (and, probably, some close relatives), a verrucomicrobe, and hence another baritone. Jenkins et al. (2002a); Schlieper et al. (2005); Michie & Löwe (2006). As the image indicates, Verrucomicrobia possess structures which might be a bit hard to explain without invoking tubulin [11]. The Prosthecobacter tubulin is so similar to eukaryotic tubulin, in fact, that it even binds a kinesin homologue, kinesin being one of the molecular shuttles that allow microtubules to be used as railway systems for transport in the eukaryotic cell. To our intense irritation, the authors of these studies always add a few unnecessary paragraphs babbling about HGT -- as if Prosthecobacter had emitted some loud evolutionary flatulence which required an apology.
 No such excuse is required. As far as we can tell, there is only one possible interpretation of this perplexing set of facts, barring arbitrary invocation of the deus ex machina of HGT. This interpretation is that the baritones, unlike all other Eubacteria, have a mode of reproduction in which FtsZ is relatively unimportant. Under some circumstances, baritones can adapt to life without using FtsZ at all. When this happens, one of two further events will occur: (a) the FtsZ gene will be lost or (b) it will be exapted to perform some other role, e.g., to form a cytoskeleton. [12] This sounds right. Many baritones (including all planctomycetes) and their tenor second cousins, the α-Proteobacteria, tend to reproduce by budding, rather than FtsZ-dependent binary fission. Angert (2005); Fuerst (1995). The recurring lack of a conventional peptidoglycan cell wall may also be relevant, since L-forms also exhibit reduced dependence on FtsZ. Onoda et al. (2000). Thus, at least some of these organisms took path a) and scrapped their FtsZ. Prosthecobacter took path b) and evolved an extremely tubulin-like protein from FtsZ.
No such excuse is required. As far as we can tell, there is only one possible interpretation of this perplexing set of facts, barring arbitrary invocation of the deus ex machina of HGT. This interpretation is that the baritones, unlike all other Eubacteria, have a mode of reproduction in which FtsZ is relatively unimportant. Under some circumstances, baritones can adapt to life without using FtsZ at all. When this happens, one of two further events will occur: (a) the FtsZ gene will be lost or (b) it will be exapted to perform some other role, e.g., to form a cytoskeleton. [12] This sounds right. Many baritones (including all planctomycetes) and their tenor second cousins, the α-Proteobacteria, tend to reproduce by budding, rather than FtsZ-dependent binary fission. Angert (2005); Fuerst (1995). The recurring lack of a conventional peptidoglycan cell wall may also be relevant, since L-forms also exhibit reduced dependence on FtsZ. Onoda et al. (2000). Thus, at least some of these organisms took path a) and scrapped their FtsZ. Prosthecobacter took path b) and evolved an extremely tubulin-like protein from FtsZ.
We can, in fact, test this hypothesis by looking for other tubulin-like structures in bugs that have lost their FtsZ -- probably not tubulins or FtsZ, but some other exaptation of this useful molecular structural element. As it turns out, planctomycetes uniquely have two or more structures which may fit this description. Anammox planctomycetes have an organelle, the anammoxosome which we will briefly discuss later), located inside a "nuclear" compartment. The anammoxosome confines the remarkably toxic intermediates (e.g. hydrazine) of the anammox reaction. The anammoxosome also displays peculiar internal tubules in thin section. Similarly, during cell division, some planctomycetes appear to have rows of tubules oriented transverse to the plane of division, apparently working at chromatin segregation. Fuerst (2005); Lindsay et al. (2001). Now look back at the peculiar Prosthecobacter-like protrusions on Pirellula staleyi, as reported by Butler et al. (2002) -- or maybe even at the image of the "crateriform structures." These last two examples are less certain, but they are suggestive and certainly fit the pattern. (Actually we have a better candidate for the crateriform structures, vide infra).
This is about as far as we can go without new data. The chain of inference is long, but each link seems to hold. Tubulin comes from FtsZ. Most Neomura have tubulin, but one group does not. Baritones are the only group of Eubacteria which (a) have tubulin, (b) don't always need FtsZ, and (c) seem to get creative with the FtsZ genes they have. This leaves the baritones as the only possible source of the Neomura.
 With respect to actin, there is much less to say. No neomuran lacks actin. No eubacterium has it. Every living organism has at least one actin homologue, one of which, in bacteria, is invariably MreB. The only hint of a phylogenetic signal we have found is the indication that MreB in stalked bacteria (yes, of course planctomycetes are stalked) may have contractile properties not found in the common run. Gitai et al. (2004). But this is a relatively vague observation. The only reason we bother to bring actin up at all is Cavalier-Smith's (2002) statement that "the origin of a cytoskeleton in a bacterium that previously had none was the key set of molecular innovations that led to phagotrophy, the endomembrane system, the nucleus and the cilium." That can't be correct. All bacteria have a reasonably elaborate cytoskeleton. See Michie & Löwe (2006) for a recent review.
With respect to actin, there is much less to say. No neomuran lacks actin. No eubacterium has it. Every living organism has at least one actin homologue, one of which, in bacteria, is invariably MreB. The only hint of a phylogenetic signal we have found is the indication that MreB in stalked bacteria (yes, of course planctomycetes are stalked) may have contractile properties not found in the common run. Gitai et al. (2004). But this is a relatively vague observation. The only reason we bother to bring actin up at all is Cavalier-Smith's (2002) statement that "the origin of a cytoskeleton in a bacterium that previously had none was the key set of molecular innovations that led to phagotrophy, the endomembrane system, the nucleus and the cilium." That can't be correct. All bacteria have a reasonably elaborate cytoskeleton. See Michie & Löwe (2006) for a recent review.
Speaking of actin, you may wonder where myosin came from -- at least, we wondered. Hopefully, you have more important issues which occupy your idle thoughts. The current understanding is that myosin is derived from the ams gene product. Ams in Escherichia is located on the mre operon, not far from mreB. Despite the usual carping about lack of sequence similarity, the N-terminal portion of Ams actually cross-reacts with antibodies directed
 against non-muscle myosins. That's good enough for us, since antibodies don't usually make the same mistake with myosin from muscles. Mcdowall et al. (1993); Okada et al. (1994). Once again, the lessons are that (a) structure trumps sequence and (b) the gap between Eubacteria and Neomura is generally smaller than one expects.
against non-muscle myosins. That's good enough for us, since antibodies don't usually make the same mistake with myosin from muscles. Mcdowall et al. (1993); Okada et al. (1994). Once again, the lessons are that (a) structure trumps sequence and (b) the gap between Eubacteria and Neomura is generally smaller than one expects.
A third lesson relates to scenarios. From a scenario-based perspective, Cavalier-Smith's concept yields a nice, orderly progression: (1) cytoskeleton → 2) loss of cell wall → (3) phagocytosis → 4) endomembranes for digestion → (5a) endoplasmic reticulum and (5b) nucleus → (6) reorganization of transcription and translation. Yet there is no evidence that this is the order in which things actually happened. If anything, the facts suggest (but do not demand) almost the reverse sequence. We'd speculate something like (2), (5b +6), 5(a), (4), (3), (1). But that's all a wild guess at this point. The point is that scenario-based, "transition" analysis rests on the subjective assignment of the probabilities for one transition over another. Sometimes, that's all we can do. However, the experience of vertebrate paleontology (where these things can, sometimes, be tested against a fossil record) is that it's better to rely on actual organisms to establish the order of trait acquisition. Accordingly, we look to Planctomycetes, the distribution of traits in the Archaea, and similar data as a guide, rather than our own assessment of what is or is not a probable transition. Finally, we should mention one model which we haven't explored, for lack of technical knowledge -- the genome of the monster virus Mimivirus, large chunks of which seem to have been derived from a very early eukaryote. Raoult et al. (2004); but c.f. Koonin et al. (2006).
Our discussion here is limited by the tendency of traditional biochemistry to speak of "bacterial" pathways as if bacteria were essentially fungible, with a few extremophile exceptions. This is far from being the case, as our discussion of lipid biosynthesis might suggest. Biochemically, bacteria are probably an order of magnitude more diverse than eukaryotes. That's one excellent reason for suspecting that Cavalier-Smith may be correct in arguing that the bacteria are paraphyletic. However, in the case of Planctomycetes, there is something to be said for the traditional view. Planctomycetes have a great many trophic styles, but they all tend to be oligotrophs. Jenkins et al. (2002). This true even of the comparatively specialized anammox planctomycetes. Strous et al. (2006). That is, planctomycetes can use almost anything as a carbon source. Rhodopirellula has the full set of enzymes necessary for fermentation to pyruvate or acetate, for oxidative metabolism, and for some (but not many) additional bells and whistles. As mentioned, it also comes equipped with a surprising number of enzymes for handling nitrogen and sulfur, with the result that it can manufacture all of the amino acids, digest chitin, and generally get by on almost nothing and almost anything, with or without oxygen. Like the planctomycete sterol synthesis pathway, all of these systems tend to be short and to the point, with no frills. Glöckner et al. (2003).
Thus, planctomycetes are the Wal-Mart of the Eubacteria. In an admittedly vague sort of way, this is what we would expect of the ancestral neomurans. Given the metabolic diversity of the Archaea, the earliest sopranos might well have had a broad metabolic repertoire. Given the metabolic homogeneity of most basal Eukarya, the soprano ancestor was probably oxygen-tolerant, if not aerobic. The baritones therefore make reasonably good candidates, although they are only somewhat better in this regard than the altos.
 Be that as it may, we know more about the weird stuff than about the diversity of ordinary metabolism and biosynthesis. Fortunately, the Planctomycetes include the metabolically unique anammox bacteria, which are strange enough to have attracted more serious attention.
Be that as it may, we know more about the weird stuff than about the diversity of ordinary metabolism and biosynthesis. Fortunately, the Planctomycetes include the metabolically unique anammox bacteria, which are strange enough to have attracted more serious attention.
These chemoautotrophic anammox planctomycetes (Brocadia, Kuenenia, Scalindua, etc.) convert nitrite and ammonia into water and nitrogen gas: NO2- + NH3+ → N2 + 2H2O. This is, obviously, a neat trick. Commercial development of planctomycetes for treating nitrogenous waste is in the engineering stages, and could save an eyebrow-raising 90% of the cost of equivalent chemical methods now in use. Op den Camp et al. (2006). Fortunately, we won't need to get into the details, since the most recent model we have seen of this reaction appears to be materially different from earlier versions (Strous et al, 2006); and there seems to be considerable uncertainty about or diversity in) the manner in which the anammox reaction is actually integrated into cell metabolism (Schouten et al., 2004).
It has been noted (i.e. we could swear we saw this, but have lost the cite) that the anammox reaction is essentially intermediate
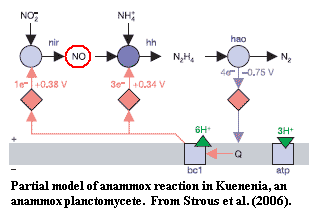 between the various types of nitrogen metabolism found in Proteobacteria and Archaea. In order to really do this right, we'd have to research more hard-core biochemistry than we want to write or you want to read. Accordingly, we've attempted to cram everything into a small diagram, based on a close reading of Strous et al (2006) and some reasonable assumptions. [14]
between the various types of nitrogen metabolism found in Proteobacteria and Archaea. In order to really do this right, we'd have to research more hard-core biochemistry than we want to write or you want to read. Accordingly, we've attempted to cram everything into a small diagram, based on a close reading of Strous et al (2006) and some reasonable assumptions. [14]
Nitrogen (N2) fixation genes are phylogenetically promiscuous. Strous doesn't mention it (so it's not on the diagram), but Kuenenia has a probable nitrogen fixation gene (nif) as well. See InterPro Q1Q2D7. The other reactions, those involved in nitrate assimilation, nitrification, and denitrification, are things that various groups in both the Proteobacteria and Archaea do, using largely homologous sets of enzymes. The usual explanations for this remarkable coincidence are HGT and/or fancy nitrogen metabolism in LUCA. Both explanations seem a little bit ad hoc -- or perhaps completely daft, depending on how honest one is being with one's self. By contrast, if we start from the anammox reactions, it isn't much of a stretch to get all three processes working properly. The anammox reaction is a typical planctomycete pathway: short, direct, inefficient, and (by Phanerozoic standards) weird. Obviously, more sensible and specialized bugs would find more reasonable and efficient ways to accomplish the desired results, thereby reaping the usual happy evolutionary rewards of efficiency and specialization. The wonder is that planctomycetes are still around for us to notice their intermediate status. Perhaps they continue to thrive only because it is nearly impossible to starve a planctomycete to death.
 Much the same pattern emerges from the C1 metabolism of planctomycetes. You may think that we have forgotten that this is supposed to be a piece on Eukarya, not Archaea. However, our thesis is the bacterial source of the Neomura, which includes both. In this scheme, the Planctomycetes would be sandwiched between tenors (Proteobacteria) and sopranos, including the archaean mezzos. The tenors are notable for their methanotrophs, the Archaea for their methanogens. That is, some Proteobacteria eat methane and spit out CO2. Some Archaea eat CO2 and spit out methane. Actually, it's nowhere near this simple, but will have to do for a first approximation.
Much the same pattern emerges from the C1 metabolism of planctomycetes. You may think that we have forgotten that this is supposed to be a piece on Eukarya, not Archaea. However, our thesis is the bacterial source of the Neomura, which includes both. In this scheme, the Planctomycetes would be sandwiched between tenors (Proteobacteria) and sopranos, including the archaean mezzos. The tenors are notable for their methanotrophs, the Archaea for their methanogens. That is, some Proteobacteria eat methane and spit out CO2. Some Archaea eat CO2 and spit out methane. Actually, it's nowhere near this simple, but will have to do for a first approximation.
As it turns out, the two sets of reactions have a large set of common steps except, of course, that the Archaea run them in one direction and the Proteobacteria in the other. They even use some of the same enzymes, together with the same cofactors. These cofactors -- methanofuran and tetrahydromethanopterin, are complex, multi-ringed structures which look a bit like a nucleotide on steroids -- which is reasonable, since they are related to both nucleotides and sterols.
Planctomycetes also share the core of this enzyme complex, including the odd cofactors, although planctomycetes don't specialize in methane metabolism any more than they specialize in anything else. Nobody outside of the methanogenic Archaea, methanotrophic Proteobacteria, and planctomycetes comes close. The planctomycetes appear to use this pathway largely to detoxify formaldehyde, a substance which may have been unpleasantly common in the Archean and early Paleoproterozoic. Chistoserdova et al. (2004). See also, Glöckner et al. (2003). By no coincidence at all, the formaldehyde step is also where methanotrophs can shuttle inorganic carbon into biosynthetic processes (Hanson & Hanson, 1996), an ability shared by planctomycetes. The planctomycete C1 enzymes are, by sequence and arrangement, equally distant from both Proteobacteria and Archaea. Chistoserdova et al. (2004). In fact, more recent work by the same group found that some methanopterin-linked genes from an environmental sample -- either planctomycetes or something entirely new -- were substantially closer to their Archaean homologues than to proteobacterial genes. Kalyuzhnaya et al. (2005).
Chistoserdova and co-workers, after presenting all sorts of dodgy HGT possibilities, ultimately confront the inescapable implications of the data and conclude as follows:
The new results presented here suggest the possibility that the methanopterin/methanofuran-linked C1 transfer pathway between the oxidation levels of formaldehyde and formate may have been an early, important function for life, and then became the first building block in the formation of both methanogenesis and methanotrophy. The functions specific to either methanogenesis or methanotrophy, such as methyl-CoM reductase, methane monooxygenase, and the specific accessory functions would have emerged later in prokaryotic history. In conclusion, the data we present here provide new insight into the history of two environmentally significant bioconversions, methanogenesis and methylotrophy, pointing to Planctomycetes, an enigmatic division of Bacteria, as potential ancestors of the key C1 transfer functions...
Finally, since HGT has achieved a vice-like stranglehold on the topic of bacterial evolution, we thought it would be worth pointing out that methane and nitrogen metabolism tend to be a package deal. It's really not clear why this should be so, but the interconnections appear to be both numerous and varied. See Hanson & Hanson (1996) for a review emphasizing classical biochemistry.So, for example, ammonia monooxygenase, which catalyzes the first step in ammonia oxidation, is closely related, by structure and function, to methane monooxygenase, which performs the first step in methane oxidation. Overall, the relationship is nebulous, if persistent. Yet it is hard to see why any relationship should exist at all if the component stepping stones on these paths are shuffled, more or less randomly, among lineages.
CONTINUED ON NEXT PAGE
ATW061129.
checked ATW061130, edited RFVS111204
 This part is much easier. You won't have to memorize a bunch of incomprehensible enzyme nicknames or vainly attempt to recall metabolic pathways you tried to memorize about six hours before the final -- and never thought about again. (Did you think we didn't know about that?) Even better, just about everyone agrees on the homologies: actin = MreB, or possibly ParM [13], and tubulin = FtsZ. We'll mention some other homologies, but those are the only two which count.
This part is much easier. You won't have to memorize a bunch of incomprehensible enzyme nicknames or vainly attempt to recall metabolic pathways you tried to memorize about six hours before the final -- and never thought about again. (Did you think we didn't know about that?) Even better, just about everyone agrees on the homologies: actin = MreB, or possibly ParM [13], and tubulin = FtsZ. We'll mention some other homologies, but those are the only two which count. FtsZ is, obviously, not functionally identical to tubulin. It has an entirely different sequence from tubulin. It does not use its GTPase in quite the same way; and it forms a associates with other strands in a different way. However, it is practically a duplicate of tubulin by structure, as the figure indicates.
FtsZ is, obviously, not functionally identical to tubulin. It has an entirely different sequence from tubulin. It does not use its GTPase in quite the same way; and it forms a associates with other strands in a different way. However, it is practically a duplicate of tubulin by structure, as the figure indicates.  No such excuse is required. As far as we can tell, there is only one possible interpretation of this perplexing set of facts, barring arbitrary invocation of the deus ex machina of HGT. This interpretation is that the baritones, unlike all other Eubacteria, have a mode of reproduction in which FtsZ is relatively unimportant. Under some circumstances, baritones can adapt to life without using FtsZ at all. When this happens, one of two further events will occur: (a) the FtsZ gene will be lost or (b) it will be exapted to perform some other role, e.g., to form a cytoskeleton.
No such excuse is required. As far as we can tell, there is only one possible interpretation of this perplexing set of facts, barring arbitrary invocation of the deus ex machina of HGT. This interpretation is that the baritones, unlike all other Eubacteria, have a mode of reproduction in which FtsZ is relatively unimportant. Under some circumstances, baritones can adapt to life without using FtsZ at all. When this happens, one of two further events will occur: (a) the FtsZ gene will be lost or (b) it will be exapted to perform some other role, e.g., to form a cytoskeleton.  With respect to actin, there is much less to say. No neomuran lacks actin. No eubacterium has it. Every living organism has at least one actin homologue, one of which, in bacteria, is invariably MreB. The only hint of a phylogenetic signal we have found is the indication that MreB in stalked bacteria (yes, of course planctomycetes are stalked) may have contractile properties not found in the common run.
With respect to actin, there is much less to say. No neomuran lacks actin. No eubacterium has it. Every living organism has at least one actin homologue, one of which, in bacteria, is invariably MreB. The only hint of a phylogenetic signal we have found is the indication that MreB in stalked bacteria (yes, of course planctomycetes are stalked) may have contractile properties not found in the common run.  against non-muscle myosins. That's good enough for us, since antibodies don't usually make the same mistake with myosin from muscles.
against non-muscle myosins. That's good enough for us, since antibodies don't usually make the same mistake with myosin from muscles.  Be that as it may, we know more about the weird stuff than about the diversity of ordinary metabolism and biosynthesis. Fortunately, the Planctomycetes include the metabolically unique anammox bacteria, which are strange enough to have attracted more serious attention.
Be that as it may, we know more about the weird stuff than about the diversity of ordinary metabolism and biosynthesis. Fortunately, the Planctomycetes include the metabolically unique anammox bacteria, which are strange enough to have attracted more serious attention. between the various types of nitrogen metabolism found in Proteobacteria and Archaea. In order to really do this right, we'd have to research more hard-core biochemistry than we want to write or you want to read. Accordingly, we've attempted to cram everything into a small diagram, based on a close reading of
between the various types of nitrogen metabolism found in Proteobacteria and Archaea. In order to really do this right, we'd have to research more hard-core biochemistry than we want to write or you want to read. Accordingly, we've attempted to cram everything into a small diagram, based on a close reading of  Much the same pattern emerges from the C1 metabolism of planctomycetes. You may think that we have forgotten that this is supposed to be a piece on Eukarya, not Archaea. However, our thesis is the bacterial source of the Neomura, which includes both. In this scheme, the Planctomycetes would be sandwiched between tenors (Proteobacteria) and sopranos, including the archaean mezzos. The tenors are notable for their methanotrophs, the Archaea for their methanogens. That is, some Proteobacteria eat methane and spit out CO2. Some Archaea eat CO2 and spit out methane. Actually, it's nowhere near this simple, but will have to do for a first approximation.
Much the same pattern emerges from the C1 metabolism of planctomycetes. You may think that we have forgotten that this is supposed to be a piece on Eukarya, not Archaea. However, our thesis is the bacterial source of the Neomura, which includes both. In this scheme, the Planctomycetes would be sandwiched between tenors (Proteobacteria) and sopranos, including the archaean mezzos. The tenors are notable for their methanotrophs, the Archaea for their methanogens. That is, some Proteobacteria eat methane and spit out CO2. Some Archaea eat CO2 and spit out methane. Actually, it's nowhere near this simple, but will have to do for a first approximation.