GroEL
Bad-Tempered Introduction
 Despite the name, GroEL is not some childhood friend of Superman. GroEL is a chaperonin. If you are unfamiliar with that term, then you need to read the glossary entry first. Return here only if you are unusually stubborn or really can't find a better source of information. In the latter case, we might tactfully suggest that you take up hopscotch, or some less intellectually demanding activity, as more suited to your research skills. This is only a brief entry designed to remind us of a few aspects of chaperonin structure and function which are relevant to our broader phylogenetic purposes.
Despite the name, GroEL is not some childhood friend of Superman. GroEL is a chaperonin. If you are unfamiliar with that term, then you need to read the glossary entry first. Return here only if you are unusually stubborn or really can't find a better source of information. In the latter case, we might tactfully suggest that you take up hopscotch, or some less intellectually demanding activity, as more suited to your research skills. This is only a brief entry designed to remind us of a few aspects of chaperonin structure and function which are relevant to our broader phylogenetic purposes.
Why Chaperonins?
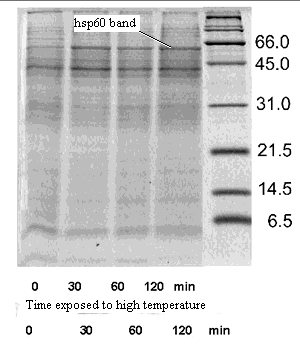
Chaperonins were originally characterized as a subpopulation of "heat-shock proteins," proteins whose synthesis was promoted by heat stress. This is the reason that GroEL and its homologues, for example, are often referred to as "Hsp60" proteins. GroEL is a 60D (actually 59D) heat shock protein.
It was realized quite early on that chaperonins help to turn polypeptides, fresh from the ribosome, into properly folded proteins. No one knew how the trick was done. In fact, the details are still obscure. However, it was clear that randomly ordered polypeptides passed through the double-ring of the chaperonin molecule and emerged as globular proteins, more or less correctly folded. The chaperonin was thus like a stage magician who runs a scarf through a magic ring, from which it emerges a different color. This illusion is normally created using a scarf that is actually a bag, with the inside covered in cloth of a different color. While feeding the "scarf" through the ring, the magician simply turns the bag inside-out. The analogy is not perfect, but we will see that the chaperonin protein does its job in roughly the same way.
But why bother? A few proteins are absolutely dependent on chaperonins to attain their correct conformation. These proteins are sometimes very important, but they represent a very small proportion of total protein diversity. Why would cells evolve general purpose chaperonins, rather than a few, special purpose types specialized to handle the bulk protein products of the cell? This generality of chaperonins became even more inexplicable when it was found that many proteins which are not chaperonin-dependent fold themselves just fine, and at about the same speed, if they are simply left to themselves in a test tube. So what is the point?
 The answer seems to relate to the fact that a cell is not a test tube. In the test tube, proteins are relatively pure and dilute. In the cell, proteins are at high concentration. Before they can fold properly, they've gotten tangled up with other things and the whole business would become a gooey mess. In fact, in many cases, the function of chaperonins may simply analogous to playpens, evolved to keep untrained infant polypeptides from blundering into other things and getting into trouble.
The answer seems to relate to the fact that a cell is not a test tube. In the test tube, proteins are relatively pure and dilute. In the cell, proteins are at high concentration. Before they can fold properly, they've gotten tangled up with other things and the whole business would become a gooey mess. In fact, in many cases, the function of chaperonins may simply analogous to playpens, evolved to keep untrained infant polypeptides from blundering into other things and getting into trouble.
In addition to folding new proteins, chaperonins also re-fold old proteins which have somehow gotten twisted out of shape. This "personal trainer" function seems to be why chaperonins are induced by heat shock. Heat stress can denature proteins, and the cell mobilizes chaperonins like a small army of physical therapists to twist everything back into its proper conformation.
 What does GroEL do?
What does GroEL do?
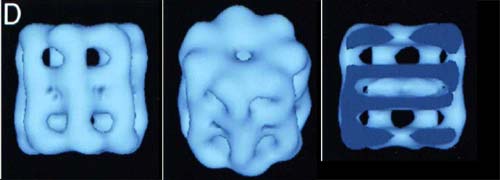
The blue-colored image above shows some low-resolution electron density maps of the holoenzyme. As the top view shows, the top half is a ring made of seven identical monomers. The bottom half is a mirror image. The third picture shows a vertical cross-section. Note the large amount of empty space in the middle of the rings.
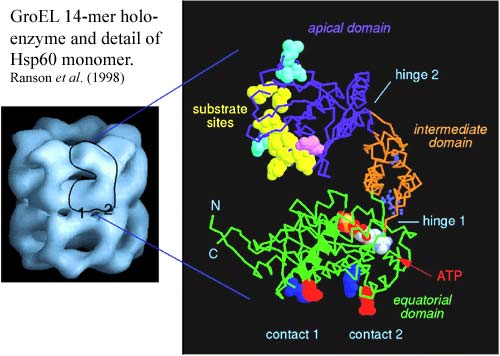
The key to the activity of GroEL is in the structure of the monomer, shown in the next image from Ranson et al. (1998). The Hsp60 monomer has three distinct sections separated by two hinge regions. The apical section contains a large number of hydrophobic binding sites for "native" (unfolded) protein substrate. Note that most sensibly folded globular proteins won't bind to the apical domain because they have their hydrophobic parts tucked away inside, away from the aqueous medium. This is the thermodynamically optimum conformation. So, these "substrate sites" will only bind to proteins which are not optimally folded. The apical domain also has binding sites for the Hsp10 monomers of its helper protein, GroES, which we'll get to in a minute.
The equatorial domain has a slot near the hinge point for ATP, as well as two attachment points for the other half of the GroEL molecule. The rest of the equatorial section is moderately hydrophilic.
What happens when we plug in ATP and GroES? The answer is shown in the next set of figures. For purposes of the following discussion, we will refer to the "activated" half of the GroEL molecule as the cis domain and the other half as the trans domain.
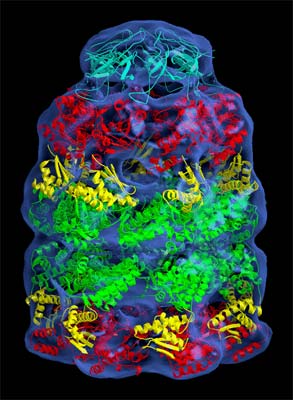
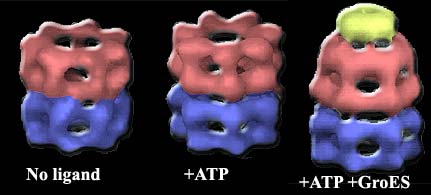 The addition of ATP and GroES has a drastic effect on the conformation of the cis domain. This effect is caused by flexion and rotation at the two hinge points on the Hsp60 monomers. The intermediate domain folds down and inward about 25° on the lower hinge. This effect, multiplied through the cooperative flexing of all monomers, increases the equatorial diameter of the GroEL cage. But the effect on the upper hinge is far more drastic. The apical domain rotates a full 60° degrees up and out on the upper hinge, and also rotates 90° around the hinge axis. This motion opens the cage very widely at the top of the cis domain, but the exit is blocked by the GroES cap, and the change in shape completely removes the substrate binding sites from the inside of the GroEL cage. [3]
The addition of ATP and GroES has a drastic effect on the conformation of the cis domain. This effect is caused by flexion and rotation at the two hinge points on the Hsp60 monomers. The intermediate domain folds down and inward about 25° on the lower hinge. This effect, multiplied through the cooperative flexing of all monomers, increases the equatorial diameter of the GroEL cage. But the effect on the upper hinge is far more drastic. The apical domain rotates a full 60° degrees up and out on the upper hinge, and also rotates 90° around the hinge axis. This motion opens the cage very widely at the top of the cis domain, but the exit is blocked by the GroES cap, and the change in shape completely removes the substrate binding sites from the inside of the GroEL cage. [3]
Once More from the Top, with Substrate
Despite all this beautiful work with empty cages, it is not clear what happens when actual proteins are provided as substrate. Plainly, the non-polar, hydrophobic interactions with the inactive state of GroEL select badly folded peptides. In effect, these confused and potentially antisocial proteins are suckered into the GroEL cage by non-polar binding sites. Then the trap is sprung, the door locked by GroES, and they find themselves caught, alone in a relatively polar environment. A second or two later, the ATP is hydrolyzed, and the peptide is pushed out through the trans domain, emerging into the cellular environment once more, but now transformed into a model molecular citizen. How has this rehabilitation taken place?
 On this topic a great deal of elegant thermodynamic and kinetic work has been done in the last 5–7 years. Most of this effort, we submit, has been wasted because of a general failure to take account of the phylogenetic framework in which this all occurs. Some thermodynamic generalities are worth emphasis. The constricted nature of the molecular cage strongly favors compact molecular conformations of the substrate protein. Free in solution, long-range, non-polar interactions can only occur at a high cost in entropy [2]. In the crowded quarters of the GroEL cage, the relative loss of entropy is much smaller. Thirumalai et al. (2003); see also Takagi et al. (2003). The method of capture also tends to concentrate the non-polar binding sites separately from the polar sites. When the GroEL non-polar surfaces are removed, the chance that any given non-polar group will encounter a nonpolar intramolecular site are much greater than in bulk solution.
On this topic a great deal of elegant thermodynamic and kinetic work has been done in the last 5–7 years. Most of this effort, we submit, has been wasted because of a general failure to take account of the phylogenetic framework in which this all occurs. Some thermodynamic generalities are worth emphasis. The constricted nature of the molecular cage strongly favors compact molecular conformations of the substrate protein. Free in solution, long-range, non-polar interactions can only occur at a high cost in entropy [2]. In the crowded quarters of the GroEL cage, the relative loss of entropy is much smaller. Thirumalai et al. (2003); see also Takagi et al. (2003). The method of capture also tends to concentrate the non-polar binding sites separately from the polar sites. When the GroEL non-polar surfaces are removed, the chance that any given non-polar group will encounter a nonpolar intramolecular site are much greater than in bulk solution.
Given these generalities, one might well speculate that the transformation works very much like the magician's ring. The hydrophobic sites on the outside of the polypeptide are gathered together at the top of the cis domain When the GroEL binding sites are suddenly tucked away, the substrates hydrophobic sites naturally bind each other. In fact, they must bind each other since the only alternative is to sit next to polar sites or to water. By definition, they don't like to do that. Conversely, the hydrophilic sites are now spread out all through the cage. Thus, they surround the clustered non-polar core as the refolded peptide is pushed out the trans side of the cage. In effect, the GroEL complex, like the magician's ring, simply operates by turning the substrate inside out.
GroEL has no Specific Mechanism
These are just general themes. The real point is that there is no need to invoke any more specific or detailed mechanism. In fact, one of the key observations made by Ranson et al. in their review is that there seems to be considerable variability in exactly when ATP and GroES bind, exactly how they effect the conformational changes, how the substrate complex forms, and how it is released.
In phylogenetic context it is easy to see why this is so. GroEL and its homologues have not changed much since the basal split between Archaea and Eubacteria. Proteins have been learning how to optimize their reactions with GroEL, or very similar molecules, for about three billion years. Given this enormous span of time, and the virtual immutability of GroES, as opposed to the huge evolutionary lability of its substrates, it is much more reasonable to conclude that cellular proteins have evolved to make the best possible uses of GroEL—not that GroEL has some magic mechanism which assures optimal results for all proteins.
GroEL is a complicated molecule and, doubtless, there are any number of ways in which a polypeptide can interact with it. Merely by way of example, it is known that certain larger polypeptides, which can't fit inside the cis chamber, extend into the trans portion and interact in some (unknown) way with the trans heptamer. Since GroEL has changed hardly at all over the last several billion years, it seems far more likely that the rest of the genome has adapted to GroEL, and uses it in various specialized ways. GroEL itself cannot specialize. GroEL, like many anatomical and biochemical features at the intersection of many developmental pathways, cannot change significantly without lethal effects. Thus the search for a specific GroEL mechanism will inevitably fail. Aside from qualitative thermodynamic considerations, such as those mentioned above, there is no evidence for a tightly constrained general mechanism, and no need to invoke one. Like the kitchen spoon, GroEL is a device which is both ubiquitous and unspecialized—not part of the recipe, but one of the basic tools around which the whole idea of a recipe is built.
[3] If you enjoy this sort of thing and have the time, take a look at Suhre et al. (2006). The authors describe free software for playing with protein conformation which comes with the specs for GroEL as a "reference case."
page uploaded ATW050124, revised ATW060902, edited RFVS111214
Text public domain. No rights reserved.
 Despite the name, GroEL is not some childhood friend of Superman. GroEL is a chaperonin. If you are unfamiliar with that term, then you need to read the glossary entry first. Return here only if you are unusually stubborn or really can't find a better source of information. In the latter case, we might tactfully suggest that you take up hopscotch, or some less intellectually demanding activity, as more suited to your research skills. This is only a brief entry designed to remind us of a few aspects of chaperonin structure and function which are relevant to our broader phylogenetic purposes.
Despite the name, GroEL is not some childhood friend of Superman. GroEL is a chaperonin. If you are unfamiliar with that term, then you need to read the glossary entry first. Return here only if you are unusually stubborn or really can't find a better source of information. In the latter case, we might tactfully suggest that you take up hopscotch, or some less intellectually demanding activity, as more suited to your research skills. This is only a brief entry designed to remind us of a few aspects of chaperonin structure and function which are relevant to our broader phylogenetic purposes.  The answer seems to relate to the fact that a cell is not a test tube. In the test tube, proteins are relatively pure and dilute. In the cell, proteins are at high concentration. Before they can fold properly, they've gotten tangled up with other things and the whole business would become a gooey mess. In fact, in many cases, the function of chaperonins may simply analogous to playpens, evolved to keep untrained infant polypeptides from blundering into other things and getting into trouble.
The answer seems to relate to the fact that a cell is not a test tube. In the test tube, proteins are relatively pure and dilute. In the cell, proteins are at high concentration. Before they can fold properly, they've gotten tangled up with other things and the whole business would become a gooey mess. In fact, in many cases, the function of chaperonins may simply analogous to playpens, evolved to keep untrained infant polypeptides from blundering into other things and getting into trouble. 
 The addition of ATP and GroES has a drastic effect on the conformation of the cis domain. This effect is caused by flexion and rotation at the two hinge points on the Hsp60 monomers. The intermediate domain folds down and inward about 25° on the lower hinge. This effect, multiplied through the cooperative flexing of all monomers, increases the equatorial diameter of the GroEL cage. But the effect on the upper hinge is far more drastic. The apical domain rotates a full 60° degrees up and out on the upper hinge, and also rotates 90° around the hinge axis. This motion opens the cage very widely at the top of the cis domain, but the exit is blocked by the GroES cap, and the change in shape completely removes the substrate binding sites from the inside of the GroEL cage.
The addition of ATP and GroES has a drastic effect on the conformation of the cis domain. This effect is caused by flexion and rotation at the two hinge points on the Hsp60 monomers. The intermediate domain folds down and inward about 25° on the lower hinge. This effect, multiplied through the cooperative flexing of all monomers, increases the equatorial diameter of the GroEL cage. But the effect on the upper hinge is far more drastic. The apical domain rotates a full 60° degrees up and out on the upper hinge, and also rotates 90° around the hinge axis. This motion opens the cage very widely at the top of the cis domain, but the exit is blocked by the GroES cap, and the change in shape completely removes the substrate binding sites from the inside of the GroEL cage.  On this topic a great deal of elegant thermodynamic and kinetic work has been done in the last 5–7 years. Most of this effort, we submit, has been wasted because of a general failure to take account of the phylogenetic framework in which this all occurs. Some thermodynamic generalities are worth emphasis. The constricted nature of the molecular cage strongly favors compact molecular conformations of the substrate protein. Free in solution, long-range, non-polar interactions can only occur at a high cost in entropy
On this topic a great deal of elegant thermodynamic and kinetic work has been done in the last 5–7 years. Most of this effort, we submit, has been wasted because of a general failure to take account of the phylogenetic framework in which this all occurs. Some thermodynamic generalities are worth emphasis. The constricted nature of the molecular cage strongly favors compact molecular conformations of the substrate protein. Free in solution, long-range, non-polar interactions can only occur at a high cost in entropy