
| Palaeos: |  |
Glossary |
| EUKARYA | Glossary P-T |
| Page Back | Unit Back | Unit Home | References | Glossary | Pieces |
| Page Next | Unit Next | Life | Dendrogram | Taxon Index | Time |
Palintomic: a palintomic cell division is a cell division without a preceding compensatory increase in size.
Parasitophorous vacuole: a vacuole within which intracellular parasites grow and develop.
Pellicle: a generic term referring to the peripheral structures which maintain cell shape and integrity. The pellicle typically includes the plasma membrane, an alveolar layer, fibrous meshes and microtubules.
Pelta: (L. pelta & Gr. πελτα = a small, light shield) in Metamonada, an anterior wall of microtubules, typically covering the nucleus. There is apparently a closely spaced series of bridges across the membrane which connect the microtubules of the axostyle and pelta. In other Protists, the term is used for any network of microtubules which forms part of the pellicle.
Peroxisome: The peroxisome is a single-membrane organelle present in nearly all eukaryotic cells. One of the most important metabolic processes of the peroxisome is the b-oxidation of long and very long chain fatty acids. The peroxisome is also involved in bile acid synthesis, cholesterol synthesis, plasmalogen synthesis, amino acid metabolism, and purine metabolism. The peroxisome for the scientist. Peroxisomes are formed by self-assembly and are not budded off from the Golgi (like lysosomes) or the endoplasmic reticulum. Peroxisomes contain oxidative enzymes, such as D-amino acid oxidase, urate oxidase, and catalase. Peroxisomes are distinguished by a crystalline structure inside a sac which also contains amorphous gray material. They are self replicating, like the mitochondria. Peroxisomes frequently function to detoxify the cell by eliminating substances like hydrogen peroxide, or other metabolites. Peroxisomes have membrane proteins that are critical for peroxisomal function, to import proteins into their interiors, proliferate or segregate to daughter cells. We had a nice picture of a peroxisome here, which was paid for by our taxes, but Florida State University didn't want you to see it. Too bad.
Phagocytosis: a method of food ingestion in which a food particle is encapsulated in a membranous food vacuole as it passes through the plasma membrane. The food vacuole then fuses with intracellular vacuoles containing digestive enzymes. See, e.g. Endocytosis.
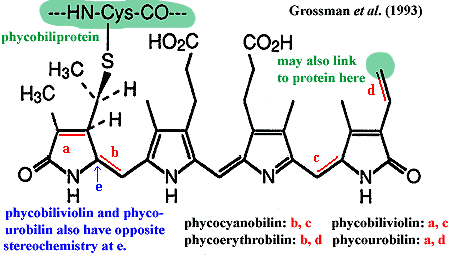 Phycobilin:
a class of light-sensitive ligands bound to
phycobiliproteins and used as accessory light-gathering pigments in algae.
The mechanism is described in the entries for phycobiliprotein
and phycobilisome. The phycobilins
are, like chlorophyll, tetrapyrrole
structures. However, unlike chlorophyll, the pyrrole rings are laid out
linearly. The detailed structure of four phycobilins commonly found in
algae are shown in the figure. The phycobilins have light absorption
maxima which vary considerably depending on their exact chemical
environment. It is also wrong to assume that any particular phycobilin
species is exclusively bound to the phycobiliprotein with a similar name.
There is considerable variation. Grossman
et al. (1993).
Phycobilin:
a class of light-sensitive ligands bound to
phycobiliproteins and used as accessory light-gathering pigments in algae.
The mechanism is described in the entries for phycobiliprotein
and phycobilisome. The phycobilins
are, like chlorophyll, tetrapyrrole
structures. However, unlike chlorophyll, the pyrrole rings are laid out
linearly. The detailed structure of four phycobilins commonly found in
algae are shown in the figure. The phycobilins have light absorption
maxima which vary considerably depending on their exact chemical
environment. It is also wrong to assume that any particular phycobilin
species is exclusively bound to the phycobiliprotein with a similar name.
There is considerable variation. Grossman
et al. (1993).
Phycobiliprotein: "water soluble fluorescent proteins derived from cyanobacteria and eukaryotic algae. In these organisms, they are used as accessory or antenna pigments for photosynthetic light collection. They absorb energy in portions of the visible spectrum that are poorly utilized by chlorophyll and, through fluorescence energy transfer, convey the energy to chlorophyll at the photosynthetic reaction center. ... The phycobiliproteins are composed of a number of subunits, each having a protein backbone to which linear tetrapyrrole chromophores are covalently bound. All phycobiliproteins contain either phycocyanobilin or phycoerythrobilin chromophores, and may also contain one of three minor bilins; phycourobilin, cryptoviolin [= phycobiliviolin?], or the 697-nm bilin. Each bilin has unique spectral characteristics, which may be further modified by interactions of the subunits and of the chromophore with the apoprotein. The phycobiliproteins in many algae are arranged in subcellular structures called phycobilisomes. These structures allow the pigments to be arranged geometrically in a manner which helps to optimize the capture of light and transfer of energy. All of the phycobiliproteins absorb incident light directly, but in addition they participate in an energy transfer chain within the phycobilisome: phycoerythrin -> phycocyanin -> allophycocyanin -> chlorophyll a." Phycobiliproteins - An Overview.
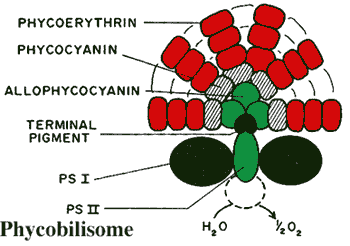 Phycobilisome:
a structure composed of several phycobiliprotein
complexes attached to the outer surface of the thylakoid
membranes in some cyanobacteria and in the chloroplasts of glaucophytes and
red algae. The phycobilisome is so arranged that a very broad range of
light frequencies can be modulated by the phycobilin pigments and the energy
transferred to chlorophyll in the light reactions of photosystem II (located
inside the thylakoid membrane) as shown in the figure. Grossman
et al. (1993).
Phycobilisome:
a structure composed of several phycobiliprotein
complexes attached to the outer surface of the thylakoid
membranes in some cyanobacteria and in the chloroplasts of glaucophytes and
red algae. The phycobilisome is so arranged that a very broad range of
light frequencies can be modulated by the phycobilin pigments and the energy
transferred to chlorophyll in the light reactions of photosystem II (located
inside the thylakoid membrane) as shown in the figure. Grossman
et al. (1993).
Piriform: pear-shaped. Often spelled pyriform.
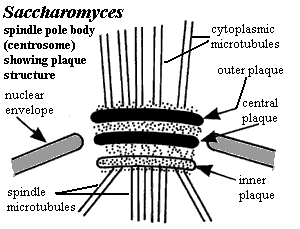 Plaque,
centrosomal: A structure present in organisms, especially Fungi, which
have a spindle pole body, rather than a conventional centrosome. The
plaque is made up of three distinct sections: outer, inner and central.
The central plaque is anchored in the plane of the nuclear envelope, the outer
plaque nucleates the cytoplasmic microtubules, and the inner plaque nucleates
the spindle microtubules in the nucleus. Yoder
et al. (2003).
Plaque,
centrosomal: A structure present in organisms, especially Fungi, which
have a spindle pole body, rather than a conventional centrosome. The
plaque is made up of three distinct sections: outer, inner and central.
The central plaque is anchored in the plane of the nuclear envelope, the outer
plaque nucleates the cytoplasmic microtubules, and the inner plaque nucleates
the spindle microtubules in the nucleus. Yoder
et al. (2003).
Plasma membrane: the principal outer membrane of the cell which encloses the cytoplasm.
plasmalemma: same as plasma membrane.
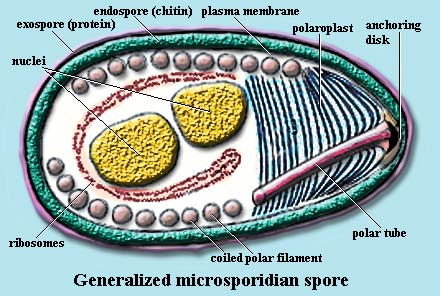 Polar
filament: in the Microsporidia,
a long, coiled series of filaments which extend the polar tube to infect host
cells.
Polar
filament: in the Microsporidia,
a long, coiled series of filaments which extend the polar tube to infect host
cells.
Polar ring: ring-shaped microtubular structures in the apical complexes of Apicomplexa. The polar rings, like most other things in the apical complex, are activated by the release of sequestered calcium ions into the parasite's cytoplasm. The presumably act in host cell invasion. The relative movement of polar rings in one system is illustrated at the glossary entry on the conoid. See also entry at subpellicular microtubules. As shown in those images, the subpellicular microtubules (if present) extend like lines of longitude from regularly spaced attachment points on the outer polar ring.
Polar tube: in the Microsporidia, the equivalent of a hypodermic needle. This organelle is inserted into a host cell and infectious sporoplasm is injected into the host.
Polaroplast: in the Microsporidia, a complex structure of layered vesicles or membranes associated with the base of the polar tube.
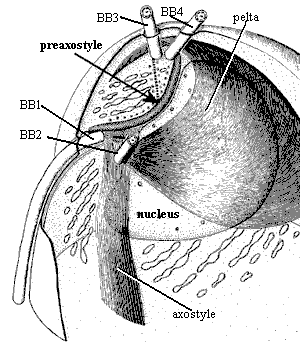 Preaxostyle: a helmet-shaped
microtubular body which caps the anterior pole of the nucleus.
Preaxostyle: a helmet-shaped
microtubular body which caps the anterior pole of the nucleus.
Pronucleus: the nucleus of a gamete, after fertilization but before complete fusion of the two haploid nuclei.
Pseudopodium: a temporary protrusion on the cell surface. Initially the cell extends a membrane process known as a lamellopodium. This is accompanied by controlled polymerization of actin filaments at the leading edge and the subsequent incorporation of these actin filaments into bundles and networks. The origin of the actual force that propels the cell forward is unknown, although it is thought to be the polymerization of the actin filaments.
Radiolaria: a taxonomic grouping approximating the Radiolaria of Haeckel (1887). It includes the Acantharea, Phaeodarea, and Polycystina, all of which are united by numerous characters such as a capsule which divides the cytoplasm into intra and extracapsular spaces, a mineralized cytoskeleton with externally-projecting spicules, numerous filipodia, and perhaps similarities of life cycle.
rDNA: the DNA which codes for ribosomal RNA. If that term isn't familiar, see the Cell Biology summary at Eubacteria. However, there are some significant differences. In contrast to prokaryotes, the four RNAs contained in eukaryotic ribosomes are coded by two types of genetic units which are generally not linked. Each type occurs in its own randomly repeated clusters. The larger unit, the rDNA, is transcribed by RNA polymerase I as a single precursor containing the small subunit (18S) rRNA, 5.8S rRNA and the large subunit (28S) rRNA, each bracketed with spacer sequences. The second type of unit codes for 5S rRNA and is transcribed by RNA polymerase III. Peyretaillade et al. (1998).
Recurrent: oriented opposite the direction of motion.
Reticulopodia: see reticulose.
Reticulose: Forming a network; characterized by a reticulated structure. Reticulose pseudopods are pseudopods in which the individual pseudopodia blend together and form irregular meshes.
 Rhoptry:
The rhoptries are club-shaped secretory organelles, often located near the apical
end of Apicomplexan intracellular parasites. Rhoptries are secreted during host
cell invasion, and rhoptry proteins are found within the lumen and the membrane
of very early stages of the forming parasitophorous
vacuole. rhoptry.html;
Sam-Yellowe
et al. (1988). Rhoptries are distinguished (somewhat) from micronemes
by the club-like shape of rhoptries.
Rhoptry:
The rhoptries are club-shaped secretory organelles, often located near the apical
end of Apicomplexan intracellular parasites. Rhoptries are secreted during host
cell invasion, and rhoptry proteins are found within the lumen and the membrane
of very early stages of the forming parasitophorous
vacuole. rhoptry.html;
Sam-Yellowe
et al. (1988). Rhoptries are distinguished (somewhat) from micronemes
by the club-like shape of rhoptries.
Ribosomal RNA: ribosomes are the small, but incredibly complex nucleoprotein complexes responsible for protein synthesis. They bind to mRNA molecules from the nucleus and physically move along the molecule, "reading" the code on the mRNA and attaching amino acids to the growing peptide (protein) chain. In eukaryotes there are three, quite distinctive RNA species bound up in the ribosome. These are known by their "Svedberg" numbers, a possibly obsolete measure of relative movement in centrifugation through a density gradient. The three species are the 5S, 18S and 28S RNAs, vide infra. Mitochondria produce their own ribosomal RNAs, the 16S and 23S rRNAs. The foolishness of using mitochondrial rRNA for phylogenetic purposes is addressed at length elsewhere.
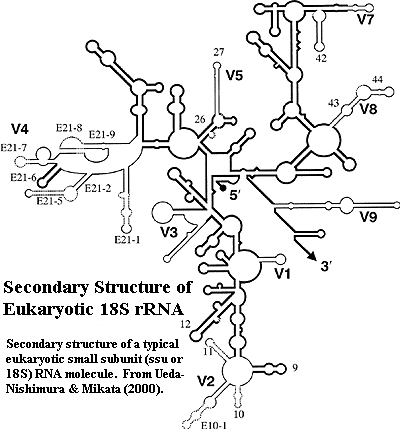 Ribosomal
RNA, 18S or ssu RNA: the RNA molecule associated with the small ribosomal subunit. The secondary structure of typical 18S rRNA is shown in the
figure from Ueda-Nishimura
& Mikata (2000). Dark regions are relatively constant. The 9
variable regions are in grey. The latter are generally referred to by the
standard nomenclature shown in the figure, vis., V1, V2,
etc. The other numbers in the figure refer to stems, but do not appear to
match the standard nomenclature for stems used by many other workers.
Ribosomal
RNA, 18S or ssu RNA: the RNA molecule associated with the small ribosomal subunit. The secondary structure of typical 18S rRNA is shown in the
figure from Ueda-Nishimura
& Mikata (2000). Dark regions are relatively constant. The 9
variable regions are in grey. The latter are generally referred to by the
standard nomenclature shown in the figure, vis., V1, V2,
etc. The other numbers in the figure refer to stems, but do not appear to
match the standard nomenclature for stems used by many other workers.
Ribosome: the cellular organelle responsible for translating mRNA into protein. Eukaryotic ribosomes are complexes of specialized RNA species and numerous proteins.
RNA polymerase: any of the enzyme complexes directly responsible for transcription -- the manufacture of RNA from the DNA template. RNA polymerase I is specialized for the synthesis of rRNA. RNA polymerase II is used to synthesize mRNAs or their precursors. RNA polymerase III is used to transcribe a single species, the 5S RNA of the large ribosomal subunit.
Rostellum: some commensal oxymonads (Oxymonadidae) have an elongate anterior structure which terminates in a holdfast, through which the cells attach to the gut wall. This is referred to as the rostellum. See image at Oxymonadidae.
rRNA: ribosomal RNA, q.v.
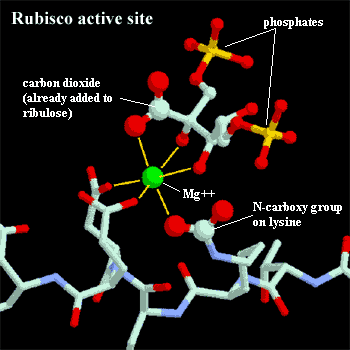 RuBisCO:
an acronym for ribulose bisphosphate carboxylase/oxygenase. Photosynthesis
is the process of fixing atmospheric carbon dioxide and transforming it into
organic carbon. RuBisCO is the enzyme which actually does the trick.
Specifically, RuBisCO attaches CO2 to ribulose bisphosphate, a five
carbon sugar. It then splits the molecule into two 3-carbon
phosphoglycerates which feed into a number of different metabolic
pathways. RuBisCO is unusually slow and inefficient. It fixes only
about three carbon dioxide molecules per second, compared to 1000+ for an
average metabolic enzyme. It is also easily confused by other substrates,
notably oxygen, and makes a remarkable number of errors. Perhaps the
evolved design, as bad as it is, can be no better. Rather than improving
the process, plants simply make enormous quantities of enzyme. RuBisCO is,
in fact, the most common protein on earth. As much as 50% of the
mass of each chloroplast is RuBisCO. Plants and algae build RuBisCO in
compact octamers, with each monomer containing two peptides. The active
site contains a magnesium ion bound by three amino acids. One of these is
a uniquely modified lysine with an extra carboxyl group added to the end of its side chain. In plant cells, this activator group, is attached to RuBisCO
during the day, turning the enzyme "on," and removed at night, turning
the enzyme "off." The exposed side of the magnesium ion binds to
both ribulose bisphosphate and the substrate carbon dioxide molecule.
RuBisCO:
an acronym for ribulose bisphosphate carboxylase/oxygenase. Photosynthesis
is the process of fixing atmospheric carbon dioxide and transforming it into
organic carbon. RuBisCO is the enzyme which actually does the trick.
Specifically, RuBisCO attaches CO2 to ribulose bisphosphate, a five
carbon sugar. It then splits the molecule into two 3-carbon
phosphoglycerates which feed into a number of different metabolic
pathways. RuBisCO is unusually slow and inefficient. It fixes only
about three carbon dioxide molecules per second, compared to 1000+ for an
average metabolic enzyme. It is also easily confused by other substrates,
notably oxygen, and makes a remarkable number of errors. Perhaps the
evolved design, as bad as it is, can be no better. Rather than improving
the process, plants simply make enormous quantities of enzyme. RuBisCO is,
in fact, the most common protein on earth. As much as 50% of the
mass of each chloroplast is RuBisCO. Plants and algae build RuBisCO in
compact octamers, with each monomer containing two peptides. The active
site contains a magnesium ion bound by three amino acids. One of these is
a uniquely modified lysine with an extra carboxyl group added to the end of its side chain. In plant cells, this activator group, is attached to RuBisCO
during the day, turning the enzyme "on," and removed at night, turning
the enzyme "off." The exposed side of the magnesium ion binds to
both ribulose bisphosphate and the substrate carbon dioxide molecule.
| Page Back | Page Top | Unit Home | Page Next |