
| Eukarya |  |
Anaeromonadida |
| METAMONADA | Oxymonadidae |
| Page Back | Unit Back | Eukarya | Eukarya References | Eukarya Dendrogram | Pieces | Taxon Index |
| Page Next | Unit Next | Unit Home | Unit References | Unit Dendrogram | Glossary | Time |
Anaeromonadida |--Trimastix `--Oxymonadida |--Polymastigidae `--+--Oxymonadidae `--+--Saccinobaculidae `--+--Pyrsonymphidae `--Streblomastigidae |
|
SummaryThis page takes up four derived groups of oxymonads, the Oxymonadidae, Saccinobaculidae, Pyrsonymphidae and Streblomastigidae. The oxymonadids are distinguished by the presence of a long proboscis-like extension at the anterior end, the rostellum. The rostellum ends in a holdfast by which the cell is fixed to the gut wall of its host. Some of the work of Guy Brugerolle on this structure is summarized below. Saccinobaculids are elongate cells, best known for their large, thick axostyles. Their unusual reproductive cycles are known from the work of LR Cleveland, some of which is also summarized here. The pyrsonymphids include two well-known genera, Pyrsonympha and Dynenympha. For many years it was thought that these were different developmental stages of the same organism. It now appears that they are distinct. Pyrsonymphids have flagella that adhere to the cell membrane and spiral around the cell, giving it a segmented appearance. The Streblomastigidae include some giant forms of the genus Streblomastix, some large enough to be seen (just barely) with out a microscope. Streblomastix is also unusual in having a genetic code which is slightly different from that used by virtually all other organisms. ATW030825. |
Oxymonadidae:
Barroella, Microrhopaladina ( = Proboscidiella = Kirbyella), Oxymonas, Sauromonas.
Range: no fossil record
Phylogeny: Oxymonadida:: (Saccinobaculidae + (Pyrsonymphidae + Streblomastigidae)) + *.
Characters: distinguished by the presence of the rostellum. Oxymonas is normally found attached in clusters to the gut wall of termites [BK97].
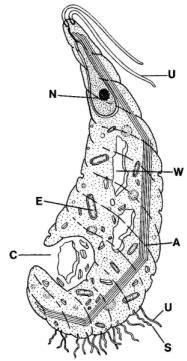
Peripheral structures: Oxymonadids have an elongated "proboscis," the rostellum, which projects anteriorly and terminates in a holdfast structure [M+03]. The rostellum can be very long -- up to 4 times the length of the cell body [BK97]. However the length is quite variable. The holdfast itself divides into finger-like projections which fix the cell to a layer of chitin in the host's gut wall [BK97]. The holdfast contains numerous longitudinally-oriented microfilaments [BK97]. The microfilaments appear to originate at the same location, at the base of the holdfast, where a system of microtubules originates. These microtubules propagate posteriorly along the length of the rostellum [BK97]. The common points of origin are small, electron-dense spots presumed to be microtubule organizing centers [BK97].
The microtubules of the rostellum are organized in convoluted ribbons which are tightly bound to a thin, fibrous sheet. The system of ribbons leaves a clear space in the center, occupied by sinuous, single microtubules. The section of the figure at right marked 'd' shows a cross-section through the rostellum illustrating this structure. The distal extent of the isolated microtubules is variable. These singlet microtubules are loosely connected by a microfibrillar matrix [BK97].
Membranes: A tuft of commensal spirochete bacteria is often found near the base of the rostellum, close to the basal bodies.
Motility organs: Oxymonadids have two pairs of flagella originating from closely associated pairs of basal bodies [K66]. These are located close to the base of the rostellum [BK97]. Microrhopaladina has two pairs of basal bodies and four flagella associated with each of its nuclei [K66]. The flagella are short, and detached cells do not appear to be motile [BK97]. The flagella of Oxymonas adhere proximally to the cell membrane, where they lie in gutters underlain by electron - dense material [KB97]. Presumably, this material is homologous to the fibers discussed by [S+02]. The two pairs of basal bodies are relatively far apart (~5µ), and are joined by the preaxostyle, to which they are fixed by microfibers.
Cytoskeleton: As usual, the preaxostyle (figure 'e' on the right) consists of a sheet of microtubules which adheres to a sheet of highly organized fibrous tissue, the whole being about 120 nm thick. The microtubular side of the preaxostyle faces the axostyle [BK97] and presumably operates as a microtubule organizing center for the latter. See image of Monocercomonoides under Oxymonadida. In Oxymonas, a second, electron-dense plate is associated with the preaxostyle, lying immediately adjacent to the plasma membrane near the base of the rostellum. An apparently unique population of spirochete bacteria bind to the cell membrane above this plate [BK97].
Many oxymonadids have two recurrent axostyle-type organs. In fact, Microrhopaladina has an axostyle associated with each of its 8-10 nuclei! [M+03]. In Oxymonas, the axostyle normally projects from the posterior end of the cell [BK97]. In cross-section (figure 'f' at right), the axostyle is a large, dense bundle of microtubules, composed of parallel stacked rows [BK97]. The microtubules in each row are tightly linked to each other. The adjacent rows also bound, but the linkage seems to be much weaker and may allow for one row to slide against its neighbors.
Most of the axostyle microtubules originate near the preaxostyle. However, the singlet microtubules from the center of the rostellum are also incorporated. The microtubular ribbons from the rostellum are shunted off to one side where they form a second axostyle-type structure of variable length [BK97].
At the moment, there is no convincing evidence that any of these structures -- rostellum, axostyle or "para-axostyle" -- is actually contractile.
Mitochondria: Oxymonadids have no mitochondria or related organelles [BK97].
Other Organelles: As with all other oxymonads, oxymonadids lack a Golgi apparatus. The ribosomes have some distinctive qualities at the molecular level. Oxymonad 18S RNA has expanded variable regions, particularly region V4. Oxymonas, and perhaps other members of the Oxymonadidae also have a similar expansion in the V7 region [M+03]. In
Nuclei: As noted, Microrhopaladina has 8 to 10 nuclei per cell [M+03], or 2-19 [K66]. In most other respects, it resembles Oxymonas [K66]. The Oxymonas nucleus is anterior [BK97]. The nucleus in Oxymonas is closely appressed to the anterior end of the axostyle [BK97].
Reproductive Cycle: The reproductive cycle of Oxymonas is similar to that of Saccinobaculus, discussed below [C56].
Image: the images of Oxymonas are from Prof. Brugerolle's website at the Laboratoire de Biologie des Protistes, Université Blaise Pascal, in Clermont-Ferrand.
Links: cytosquelette caractere.
References: Brugerolle & Konig (1997) [BK97]; Cleveland (1956) [C56]; Kudo (1966) [K66]; Moriya et al. (2003) [M+03]. ATW030823.
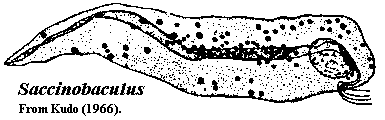 Saccinobaculidae:
Notila, Saccinobaculus.
Saccinobaculidae:
Notila, Saccinobaculus.
Range: no fossil record
Phylogeny: Oxymonadida::: (Pyrsonymphidae + Streblomastigidae) + *.
Characters: Saccinobaculids are usually elongated, but plastic cells found in the gut of wood-eating cockroaches [K66].
Peripheral structures: Saccinobaculus is reported to have a small holdfast organ, but is generally free-swimming [M+03].
Motility organs: The 4, 8 or 12 flagella adhere to the cell, but project freely [K66].
Cytoskeleton: The saccinobaculid axostyle is very large and paddle-like [K66]. It undulates and is probably involved in motility. In Saccinobaculus, the posterior end of the axostyle is enclosed in a sheath [K66].
Nuclei: The normal vegetative form is haploid in Saccinobaculus [C56]. Notila has at least two geographical variants, one diploid, and the other tetraploid [C56].
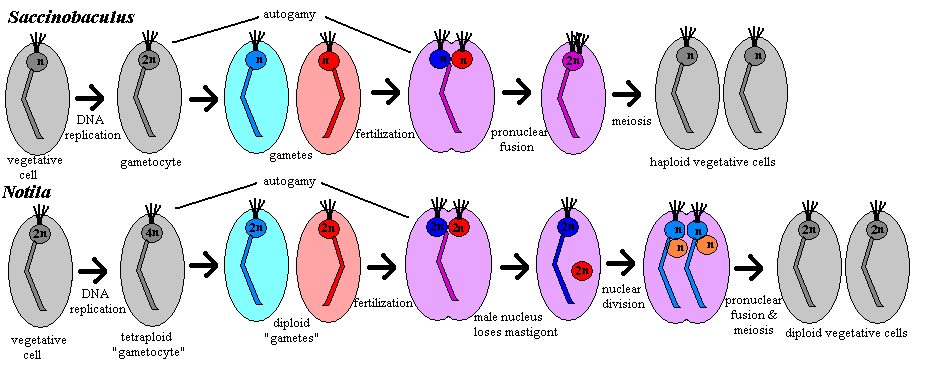
Sexual reproduction: The cycles of Notila and Saccinobaculus are rather different. In Saccinobaculus, the haploid vegetative cell undergoes a round of DNA replication during molt of the host organism, resulting in a diploid gametocyte [C56]. The gametocyte divides by mitosis, yielding two haploid gametes [C56]. During this division, as in mitosis in the vegetative cell, the axostyle is the only cell organelle discarded [C56]. In the majority of cases, the cell division is never completed, and the two nuclei simply fuse again (autogamy), forming another 2n cell with the same gene complement as the gametocyte. However, heterologous fertilization also occurs. The gametes' axostyles begin to fuse shortly before the fusion of the pronuclei [C56]. Just after pronuclear fusion begins, the process stalls, and the zygote remains in this partially fused state until ecdysis of the host insect -- 30 or 40 days under natural conditions [C56]. During this time, the zygote retains all of the organelles of both parent cells: 2 axostyles, 8 flagella and 4 centrioles [C56]. At ecdysis, 4 flagella and 2 centrioles are lost. Meiosis is then completed with a single division to create two vegetative haploid daughter cells [C56].
In Notila, the gametocyte is tetraploid. If gametogenesis is complete, this results in two diploid "gametes" [C56]. (As Cleveland notes, none of the usual nomenclature really fits the case of Notila, which is absolutely unique.) The parental axostyle is discarded and each "gamete" produces a new axostyle associated with its nucleus. The parental flagella are retained and 4 new flagella re produced. The gametes then fuse with each other (autogamy) or with a gamete from another cell (fertilization). Whether autogamy or fertilization occurs, the result is a tetraploid cell -- one hesitates to call it a zygote -- with two nuclei. One of the nuclei, the "male" for lack of a better term, loses its flagella and axostyle. Both nuclei then undergo meiosis. In the process, the daughter "female" nuclei discard their old axostyle and create a new one each, and again retain their flagella but create two additional flagella each. The result is one cell with four haploid nuclei, two with karyomastigont and two without. Each "male" nucleus then fuses with a "female" nucleus, and the cell divides again, resulting in two diploid vegetative cells [C56].
References: Cleveland (1956) [C56]; Kudo (1966) [K66]; Moriya et al. (2003) [M+03]. ATW030815.
 Pyrsonymphidae: Dinenympha,
Pyrsonympha. There has been considerable controversy concerning whether these
are two life stages of the same organism. However, Dacks et al.
[D+01] have performed an elegant series of in situ hybridization studies
which appear to establish beyond question that the two are closely related, but
distinct genera.
Pyrsonymphidae: Dinenympha,
Pyrsonympha. There has been considerable controversy concerning whether these
are two life stages of the same organism. However, Dacks et al.
[D+01] have performed an elegant series of in situ hybridization studies
which appear to establish beyond question that the two are closely related, but
distinct genera.
Range: no fossil record. Both genera are found on both sides of the Pacific in association with termites of the family Rhinotermatidae. This termite family is believed to have evolved in the Eocene [M+03].
Phylogeny: Oxymonadida:::: Streblomastigidae + *.
Characters: The pyrsonymphids are heterotrophic protists that have been found only in the hindgut of wood-eating cockroaches and termites. Many bacteria including spirochetes, can be associated with pyrsonymphids as epi- and endosymbionts. Pyrsonympha is large (170µ) and piriform [HC70]. Dinenympha is much smaller (25-50µ) and has an odd, twisted appearance like a spirochete bacterium [HC70].
Peripheral structures: Pyrsonympha bears a holdfast; however a rostellum is absent [BK97]. Dinenympha does not have a holdfast. [M+03].
Motility organs: 4 or 8 flagella and a corresponding number of basal bodies [HC70]. The flagella are all recurrent [B91]. They adhere to the cell membrane, spiraling around the outside of the cell and giving it a banded or segmented appearance [K66]. Striated fibers line the axoneme. Presumably this is another homologue of the I fiber of [S+02].
Cytoskeleton: The axostyle is motile and extends the full length of the cell. Dinenympha and Pyrsonympha have 1 and 2 preaxostyles, respectively.
Mitochondria: no mitochondria or related organs
Other Organelles: Moriya et al. [M+03] have recently published a particularly elegant series of experiments involving in situ hybridization of fluorescently-labelled 18S rDNA probes with mixed preparations from termite hindguts. The results seem to have finally put to rest the issue of whether Dinenympha and Pyrsonympha are actually different organisms. None of their probes cross-hybridized between the two genera, indicating that Dinenympha and Pyrsonympha are entirely distinct taxa. In fact, the results suggested that there may be considerable cryptic speciation within Pyrsonympha which has not been detected by morphological observations. Nevertheless, the two pyrsonymphids were both found to be valid taxa, and to be sister clades.
Nuclei: The nucleus is anterior and tends to be large [K66].
Reproduction: reproduction is timed to coincide with molting in the host termite, presumably stimulated by the same ecdysone release which serves as the host's molting signal [HC70]. A few days before the molt begins, the pyrsonymphid cells begin to undergo several rounds of palintomic division, resulting in a population of extremely small daughter cells (12-30µ) [HC70]. These are lost at molt and ingested by other termites [HC70]. Hollande & Carruette-Valentin [HC70] describe many additional details of the reproductive cycle. However their experimental system (termites in petri dishes feeding on moistened filter paper) is so far removed from natural conditions that it is difficult to be sure that they are observing a natural process. Given that their ultimate conclusion (that the two pyrsonymphid genera are different developmental stages of the same organism) appears to have been mistaken, we suspect that the process they observed is pathological.
Links: Untitled Document; Zoomastigophora- Tetramastigota (Japanese).
References: Brugerolle (1991) [B91]; Brugerolle & Konig (1997) [BK97]; Dacks et al. (2001) [D+01]; Hollande & Carruette-Valentin (1970) [HC70]; Kudo (1966) [K66]; Moriya et al. (2003) [M+03], Simpson et al. (2002) [S+02]. ATW030815.
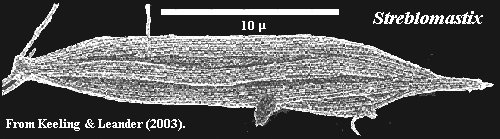 Streblomastigidae: Streblomastix.
Streblomastigidae: Streblomastix.
Range: no fossil record
Phylogeny: Oxymonadida:::: Pyrsonymphidae + *.
[KL03] performed a number of sequence comparisons within a natural population, including α-tubulin, elongation factor 1α, β-tubulin, and heat shock protein 90. In each case, the coding was virtually identical, although there was considerable variability in synonymous positions. All proteins were also close in sequence to the corresponding proteins in Pyrsonympha and Dinenympha, as well as reasonably close to Trimastix [KL03]. [M+03] assert, on the basis of similar results, that Streblomastix ought to be classified as a pyrsonymphid. However, no one disputes that the two pyrsonymphid genera are sisters. Therefore, this reduces to a pointless argument about taxonomic rank.
Characters: found only in animal gut [KL03]. This group includes giant forms of Streblomastix, measuring over 500µ, which are the largest known oxymonads [M+03].
Peripheral structures: Streblomastix bears a holdfast; however a rostellum is absent. [BK97] [M+03].
Cytoskeleton: Streblomastix and Pyrsonympha show a close relationship in an α-tubulin phylogeny [KL03].
Streblomastix has a variant genetic code in which the "universal" stop codons UAA and UAG encode glutamine [KL03]. This particular variation from the code is also found within ciliates, where it may have evolved more than once, and in some green algae and in hexamitid diplomonads. [KL03]. The variant code is not shared by Pyrsonymphids. It is likely that these are all independent departures from the standard code. UAA and UAG are not known to have ever been reassigned to any amino acid other than glutamine, so it is presumed that there may be some special affinity. That affinity may have some connection with the fact that eukaryotes (and Archaea) use the same translation termination factor to recognize all three stop codons (there are two such factors in Eubacteria). Potential for ambiguity also arises at the tRNA charging step. In eukaryotes, tRNAgln can be charged either by a specific gln-tRNA synthetase, or by the glu-tRNA synthetase, with subsequent derivatization of the amino acid by an amidotransferase (this is the only pathway to tRNAgln in Bacteria and Archaea). Thus an aberrant gln-tRNA synthetase is not necessarily fatal [KL03].
The sequence of Streblomastix small subunit ribosomal RNA (ssuRNA) contains numerous large insertions, some of which are shared by Pyrsonympha [KL03].
References: Brugerolle & Konig (1997) [BK97]; Keeling & Leander (2003) [KL03]; Moriya et al. (2003) [M+03]. ATW030815.
| Page Back | Page Top | Unit Home | Page Next |