Hox Genes - 2
But Why? ... Some Unanswered Questions
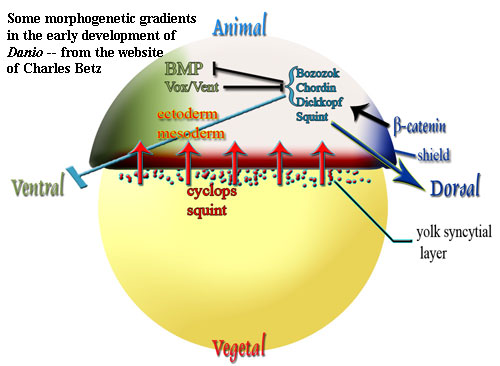 That's the idealized version. Real organisms, like real military organizations and real computer programs, tend to operate in ways which deviate from their original designs. So, before getting into specifics, we might look a little harder at the biochemical context. First, what exactly is a homeodomain anyway? Fortunately, we don't have to explain this. Pharyngula has a nice short discussion of the homeodomain. He uses language more efficiently than we do, so we will refer you to his discussion -- in particular the "Homeobox and Homeodomain" heading.
That's the idealized version. Real organisms, like real military organizations and real computer programs, tend to operate in ways which deviate from their original designs. So, before getting into specifics, we might look a little harder at the biochemical context. First, what exactly is a homeodomain anyway? Fortunately, we don't have to explain this. Pharyngula has a nice short discussion of the homeodomain. He uses language more efficiently than we do, so we will refer you to his discussion -- in particular the "Homeobox and Homeodomain" heading.
Hox genes are but one family of what Carroll (2005) aptly calls toolkit genes. These all code for relatively small, simple proteins with a DNA-binding domain (homeodomain, "zinc finger," or something more exotic) which function as messengers during development to control the transcription of whole batteries of genes. The reason Carroll uses the toolkit analogy is that these transcription factors are used to regulate different groups of genes in different organisms. He comes close to arguing that toolkit genes are completely general signals which have no inherent information content.
Carroll doesn't actually go this far, because there is a really odd consistency in the functional role these genes play. The best-known example is perhaps the non-hox homeodomain protein pax-6. Pax-6 regulates entirely different sets of genes in chordates and arthropods. However, in both cases, it is plays a critical role in eye formation. The same applies to tinman and heart development. For that matter, the hox genes have some of the same peculiarity in that they rarely change order [1] It's easy to "explain" all this consistency by assuming that all these functional associations are simply inherited from the last common ancestor of cows and crustaceans ("Urbilateria"). That's exactly the argument we made elsewhere, and that's probably all there is to it. But, if we accept that argument, it
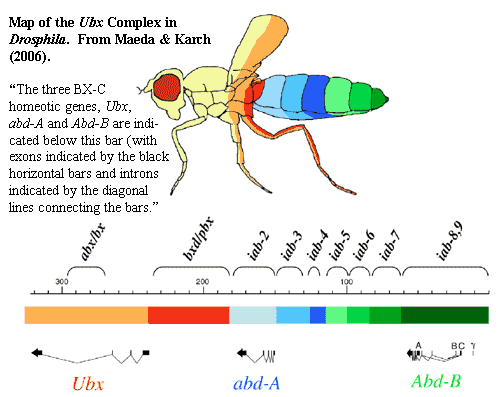 would mean that our last common Great-Grandmother with clams and shrimp was a lot more complex than is generally believed. Thus, we may shortly have to choose between the received teachings of biochemistry and phylogeny. That is, either Urbilateria was very complex and some morphologically simple bilaterian animals are probably outside the crown group Bilateria, or the Hox genes have a really peculiar biochemical link to specific body functions. This could go either way, but our current suspicion is that the guys in the lab coats are going to win, and we're going to have to adjust our phylogeny. See Ryan et al. 2007) (highly evolved anthozoan homeodomain system, discussed later), Chen et al. 2004)* (Vernanimalcula: in our own, distorted view, a possible stem group bilaterian [2]).
would mean that our last common Great-Grandmother with clams and shrimp was a lot more complex than is generally believed. Thus, we may shortly have to choose between the received teachings of biochemistry and phylogeny. That is, either Urbilateria was very complex and some morphologically simple bilaterian animals are probably outside the crown group Bilateria, or the Hox genes have a really peculiar biochemical link to specific body functions. This could go either way, but our current suspicion is that the guys in the lab coats are going to win, and we're going to have to adjust our phylogeny. See Ryan et al. 2007) (highly evolved anthozoan homeodomain system, discussed later), Chen et al. 2004)* (Vernanimalcula: in our own, distorted view, a possible stem group bilaterian [2]).
In this same vein, let's ask a more fundamental question. Why bother with this elegant, but recondite, system? Why didn't evolution just come up with a simple gradient of front to back, based on (for example) the progress of gastrulation? Morphogenetic gradient fields are very old stuff. See review by de Robertis et al. (1991). Alternatively, why require the embryo to do all this hard work in the first place, when mom could easily set up the pattern during egg development? A maternally, established gradient seems to be the way it's done in some cnidarians. Momose & Houliston (2007). In fact, that's arguably how anteroposterior "patterning" is actually accomplished in Drosophila itself
-- with a maternal bicoid gradient, before the embryo expresses hox. [12: update] In fact, in baby Drosophila, Mom even takes care of segment formation. The embryo later has to override the maternal "parasegments" in order to set up its own segmentation pattern. Maeda & Karch (2006).
Why bother? We have no answer for this question, and we will return to various aspects of the problem repeatedly. We thought perhaps that evolution of the hox system had something to do with segmentation[3]. However, bilaterian segmentation post-dates Urbilateria and was independently invented at least three times. Seaver (2003)* discussed elsewhere). Another bad idea was that the regulation of hox genes was somehow unique and important to the properties and evolution of the system. In fact, it turns out that the regulation of hox expression is quite complex, variable and -- most importantly -- it didn't immediately suggest any easy turns of arm-waving rhetoric with which to ornament this discussion. See Maeda & Karch (2006)* for a far more determined, more well-intentioned, and almost successful, attempt to make this subject comprehensible. In addition, hox genes are apparently subject to multiple levels of regulation, including post-transcriptional regulation, like any other gene. Pearson et al. (2005).
 So what, exactly, does the hox system regulate and how? Again, understanding of this area has not yet reached the point at which a few sentences would suffice to summarize a fundamental principle. We're still at a point where some individual cases are understood, but we don't have a handle on any common mechanisms tying them together.
So what, exactly, does the hox system regulate and how? Again, understanding of this area has not yet reached the point at which a few sentences would suffice to summarize a fundamental principle. We're still at a point where some individual cases are understood, but we don't have a handle on any common mechanisms tying them together.
The current state of the art is described by Pearson et al. (2005). As these folks point out, one of the relatively few strong insights gained so far is that hox proteins bind to DNA as heterodimers or heterotrimers, typically cooperating with transcription factors of the pbx or hth classes (both homeodomain-containing groups). These multi-protein complexes have different sequence specificities than a simple linear combination of the individual monomers, and this creates the potential for a relatively complex, fine-tuned system of regulation based on a small number of very simple proteins.
 Even with this expanded set of regulatory possibilities, we would expect that the hox system would work on a relatively high level. That is, its anticipated targets would be other regulatory genes rather than genes coding for individual enzymes and structural proteins. As it turns out, this is probably the case. Hox proteins, as expected, control the expression of a number of important transcription factors, such as decapentaplegic (dpp, governing formation of viscera), twist (mesoderm) and distalless (dll, appendages). Pearson et al. argue that hox signals also directly regulate lower-level "blue collar" genes. However, their examples are not particularly convincing.
Even with this expanded set of regulatory possibilities, we would expect that the hox system would work on a relatively high level. That is, its anticipated targets would be other regulatory genes rather than genes coding for individual enzymes and structural proteins. As it turns out, this is probably the case. Hox proteins, as expected, control the expression of a number of important transcription factors, such as decapentaplegic (dpp, governing formation of viscera), twist (mesoderm) and distalless (dll, appendages). Pearson et al. argue that hox signals also directly regulate lower-level "blue collar" genes. However, their examples are not particularly convincing.
But enough of vague generalities. Let's turn to ... uh ... more specific generalities. Where do hox genes come from? The basic helix-turn-helix DNA-binding motif is about as old as DNA itself. Nelson & Cox (2005); Giraldo & Díaz-Orejas 2001). The homeodomain itself is essentially identical to the DNA-binding regions of some transcriptional regulators in bacteria. Carroll (2005). While irrelevant to the present discussion, we can't resist mentioning that plants have a system somewhat analogous to the hox system involving "MADS-box" genes, which are also descended from bacterial transcription factors. Kofuji et al. 2003). In fact, plants have some homeobox genes and animals use a few MADS genes.
 Sponges and non-anthozoan Cnidaria (including Ctenophora) lack identifiable hox homologues, although they do possess closely-related homeobox genes. However the key property of colinearity is present only in anthozoans and bilaterians. Peterson et al. 2005). Despite speculations about hox gene clustering basal to the Cnidaria (e.g. Monteiro & Ferrier, 2006), "true" hox homologues are found only in certain cnidarians and in Bilateria. Brooke & Holland 2003). And, even in Anthozoa, their colinearity is not strong or consistent.
Sponges and non-anthozoan Cnidaria (including Ctenophora) lack identifiable hox homologues, although they do possess closely-related homeobox genes. However the key property of colinearity is present only in anthozoans and bilaterians. Peterson et al. 2005). Despite speculations about hox gene clustering basal to the Cnidaria (e.g. Monteiro & Ferrier, 2006), "true" hox homologues are found only in certain cnidarians and in Bilateria. Brooke & Holland 2003). And, even in Anthozoa, their colinearity is not strong or consistent.
That brings us up to Ryan et al. (2007)* who discuss the homeobox genes of the anthozoan, Nematostella, a sea anemone. This is probably the most interesting paper we've read in the last year. Fortunately it is readily available on line. We will only deal with the portion which relates to hox genes [4]. Nematostella has a surprising number of homeobox genes -- about 130. Ryan et al. (2006). This is somewhat more than in Drosophila, which has around 100. Id. Of these 130, Nematostella has seven hox genes: two hox1 orthologues, three hox2 orthologues, and two posterior class hox genes. Ryan et al. (2007). There are no hox3 or middle class homologues.
Are the Nematostella hox genes collinear? Sort of. Maybe. First, notice that, going down the second column in the figure, the sequence anthox6-anthox8a/8b-anthox1 makes a sort of anterior to posterior pattern -- at least "in the dusk, with the light behind her." More convincingly, going down the first column, the sequence anthox6a-anthox7-anthox8a-anthox8b-anthox1a makes a very good pattern along a "secondary," "dorsoventral," or (as we see it) radial axis. So, we seem to have some morphological colinearity. It's just not the kind we're used to seeing.
Genetic colinearity is a closer case. The Nematostella hox genes are fairly scattered, but there is one major exception. All of the hox2 homologues and one of the hox1s (anthox6) are tightly linked in a single cluster with several other homeodomain genes. We've run out of room here, so the image is included in footnote [5]. All of these genes are transcribed in the same direction except Dmbxd and HlxB9.
Although Ryan et al. don't go into it, the structure of this group is interesting. The hox genes are somewhat out of order, with the hox1 homologue positioned after the tightly-linked hox2 group. Dmbxd is a member of the dmbx class. This class is only found in cnidarians and deuterostomes. Morphologically, it is expressed at the anterior limit of hox expression. Takahashi & Holland (2004). Ryan & Co. don't test for expression of this gene, but it is genetically located in the right place, just upstream of the hox cluster. Evx is an even-skipped gene. Evx class genes, in bilaterians, are located just downstream of the hox cluster, often linked with a caudal element. Copf et al. (2003); Minguillón & Garcia-Fernandèz (2003); Minguillón et al. (2005). Here, the evx is downstream of the hox2 cluster, between them and the hox1 homologue.
| Phylogeny of Selected Homeodomain Gene Families |
| ■─┬─Dlx
└─┬─Hlx
├─┬─Even skipped (Evx)
│ └─Rough (ro)
└─┬─Mox
└─┬─Gsx*
└─┬─Anterior Hox
│ ├─Hox1
│ └─Hox2
└─┬─┬─┬─Hox3
│ │ └─Xlox*
│ └─Middle Hox
└─┬─Posterior Hox
└─Caudal (Cdx*) |
| Adapted from Ryan et al. (2007). The tree was constructed with a neighbor-joining technique, using homologues from Branchiostoma (a deuterostome), Drosophila (a protostome), and Nematostella. Each species gives the same pattern. Families marked '*' are ParaHox genes (explained below). |
We've droned on Nematostella and about this rather marginal hox system because it seems to be the most basal system which has any significant colinearity and is known in some detail. Potentially, it tells us a lot about colinearity, one of the central mysteries of hox, and about the evolution of the system. In addition to colinearity, Nematostella illustrates several features of the hox system which we will see repeatedly, to wit:
1. Duplication: Hox genes have a strong tendency to duplicate, often in groups (or so it is said). As Ryan y sus compadres discuss, the tendency is to assume multi-gene duplication events. However, their data suggest that the "hox cluster" system developed by single-gene duplications and gradual rearrangement. Certain Evil Norwegians disagree. However, since they seldom publish in open-access journals, we will spitefully refuse to cite them.
2. Independence of morphological and genetic colinearity: Although the Nematostella hox system has some degree of both morphological and genetic colinearity, these features show considerable independence. The posterior (Anthox1 and 1a) genes are not genetically linked to the others, but appear to work as part of a single morphological system. One hox1, and all hox2, homologues are collinear, but the hox1 gene is in the wrong order -- actually located after a gene (evx) which is usually the downstream bookend for the whole system.
3. Recycling: as the third column in the figure indicates, some hox genes are used again later in development, apparently for completely different purposes.
4. Evolutionary order of classes: Anterior class genes came first, followed by posterior class genes. The consensus view is that these were followed, later in phylogeny, by the middle class and hox3 (upper middle class) homology groups (but see phylogeny box).
We leave the Cnidaria with an unanswered question. The homeodomain genes of Nematostella include several hints that the Cnidaria are paraphyletic. In fact, some features even suggest that the Anthozoa may be more closely related to deuterostomes than to protostomes, although that seems unlikely. Other work connected with Mark Martindale Univ. Hawaii), his former student John Finnerty Boston Univ.), and Finnerty's students (e.g., Ryan) indicates that Anthozoa are morphologically and genetically very bilaterian. See, for example, Martindale et al. 2004); Sullivan et al. (2006). Yet none of this work actually questions the monophyly of the Cnidaria. Perhaps that assumption needs to be re-examined.
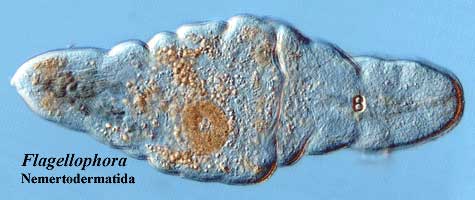 The acoelomorphs are gutless little flatworms which used to be thrown in with "Platyhelminthes" for lack of a better place to put them. That is, they were two of the many classes in the garbage taxon Platyhelminthes: Acoela and Nemertodermatida. After Platyhelminthes fell apart, these orphans were shuffled into an indifferent foster home between Urbilateria and the base of the protostomes, while the Authorities decided what to do with them. Recently, they seem to have been evicted from Bilateria altogether and live on the streets, as it were, unafilliated with any larger Metazoan group. Cook et al. (2004); Jiménez-Guri et al. (2006) (collectively, the "Barcelona Group").
[6]
The acoelomorphs are gutless little flatworms which used to be thrown in with "Platyhelminthes" for lack of a better place to put them. That is, they were two of the many classes in the garbage taxon Platyhelminthes: Acoela and Nemertodermatida. After Platyhelminthes fell apart, these orphans were shuffled into an indifferent foster home between Urbilateria and the base of the protostomes, while the Authorities decided what to do with them. Recently, they seem to have been evicted from Bilateria altogether and live on the streets, as it were, unafilliated with any larger Metazoan group. Cook et al. (2004); Jiménez-Guri et al. (2006) (collectively, the "Barcelona Group").
[6]
The Barcelona Group previously (2004) found that the acoel Symsagittifera had anterior, middle, and posterior hox genes. It even had a caudal cdx) homologue -- but lacked a hox3 class gene. This was a little odd, for reasons we'll get to in a moment. Consequently, they decided to check these results with a representative of another group of acoelomorphs, Nemertoderma. As a result of this work, they have changed their position and state that this group ancestrally had representatives of all four hox homology groups. This conclusion follows from their (2006) finding that the acoelomorph Nemertoderma has -- not a hox3 gene, but an xlox homologue.
 Sadly, this forces us to get into the ParaHox genes, a task we had hoped to avoid. Our melancholy is due to: (a) our minimal understanding of ParaHox and (b) a depressing suspicion that the underlying theory is a gross and misleading oversimplification. As we lack any inclination to get creative here, we will fall back on the mythic language of reviews and textbooks. To us, this stuff sounds a bit like a Bronze Age creation myth:
Sadly, this forces us to get into the ParaHox genes, a task we had hoped to avoid. Our melancholy is due to: (a) our minimal understanding of ParaHox and (b) a depressing suspicion that the underlying theory is a gross and misleading oversimplification. As we lack any inclination to get creative here, we will fall back on the mythic language of reviews and textbooks. To us, this stuff sounds a bit like a Bronze Age creation myth:
So it came to pass that a "... ProtoHox cluster of four genes duplicated early in animal evolution, giving rise to two twin clusters. These would be the primordial Hox cluster, which expanded by cis duplication to eight genes in Drosophila, or to 13 paralogous groups in mammals, and the primordial ParaHox cluster, which lost one member and gave rise to the three-gene complex maintained at least in cephalochordates and vertebrates." [García-Fernàndez, 2005]. So that the Hox grew and became many and multiplied in the eagles of the great hills, in all the beasts of the earth, and also among the fishes of the ocean depths did they flourish.
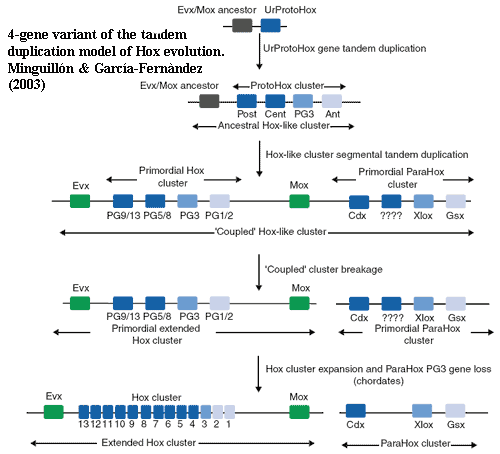
This parody is shamefully unfair; and our admittedly rudimentary) sense of fair play compels us to include a detailed graph of the actual model from Minguillón & García-Fernàndez (2003). The idea is that the hox and parahox clusters developed from tandem duplication of a four gene protohox cluster and its downstream evx book-end. When the double cluster split, it split unevenly. This left evx book-ends on both sides of the hox cluster. The upstream homologue subsequently diverged to become a mox gene.
Why come up with this messy and complex model? Look back at the hox phylogeny from Ryan et al. (2007). The hox3 and posterior homology classes are both more closely related to non-hox (parahox) genes than to each other. In the Barcelona Group's experiments, this is also true of the anterior class (paired with gsx). Also, the two book-end genes, evx and mox, are closely related. Something has to explain this odd connection. The parahox model does that. If this is correct, the presence of a cdx gene in Acoelomorpha without either a hox3 or an xlox was peculiar.
 But here's an alternative which we just invented -- don't take it too seriously). Suppose there was never a protohox cluster. Instead, morphological colinearity came first. This made it convenient, for purposes of gene regulation, to evolve genetic colinearity. Not essential, mind you. Just helpful. What kind of genes would get recruited to this new linkage group? Clearly, if we are integrating multiple complex regulatory domains, it might be helpful if we were using duplicate spare parts. In that case, while one copy was gradually fitting itself into the complex regulatory assembly line of a hox linkage group, the other copy would still be out there doing things the old-fashioned way. That is, evolution would favor recently duplicated genes, since an embryo could often survive the sub-optimal regulation of one copy.
But here's an alternative which we just invented -- don't take it too seriously). Suppose there was never a protohox cluster. Instead, morphological colinearity came first. This made it convenient, for purposes of gene regulation, to evolve genetic colinearity. Not essential, mind you. Just helpful. What kind of genes would get recruited to this new linkage group? Clearly, if we are integrating multiple complex regulatory domains, it might be helpful if we were using duplicate spare parts. In that case, while one copy was gradually fitting itself into the complex regulatory assembly line of a hox linkage group, the other copy would still be out there doing things the old-fashioned way. That is, evolution would favor recently duplicated genes, since an embryo could often survive the sub-optimal regulation of one copy.
That doesn't explain the mox/evx bookends, but it wouldn't be hard to come up with some similarly ad hox explanations for them. We can imagine several reasons why two similar genes might tend to bracket the hox cluster. However, we will not waste your time with these imaginings. The real point is that we can arrive at the same result, with the sub-families of hox genes each tending to be paired with a non-hox gene, as well as bookends, etc. without invoking tandem duplication events. Clearly, the data from Ryan et al. (2007) support this kind of sloppy, inelegant solution. Perhaps that's the answer. We are inclined to think that this is still too simple. Colinearity is, as we will see, a complex topic.
CONTINUED ON NEXT PAGE
 That's the idealized version. Real organisms, like real military organizations and real computer programs, tend to operate in ways which deviate from their original designs. So, before getting into specifics, we might look a little harder at the biochemical context. First, what exactly is a homeodomain anyway? Fortunately, we don't have to explain this. Pharyngula has a nice short discussion of the homeodomain. He uses language more efficiently than we do, so we will refer you to his discussion -- in particular the "Homeobox and Homeodomain" heading.
That's the idealized version. Real organisms, like real military organizations and real computer programs, tend to operate in ways which deviate from their original designs. So, before getting into specifics, we might look a little harder at the biochemical context. First, what exactly is a homeodomain anyway? Fortunately, we don't have to explain this. Pharyngula has a nice short discussion of the homeodomain. He uses language more efficiently than we do, so we will refer you to his discussion -- in particular the "Homeobox and Homeodomain" heading.  would mean that our last common Great-Grandmother with
would mean that our last common Great-Grandmother with  So what, exactly, does the hox system regulate and how? Again, understanding of this area has not yet reached the point at which a few sentences would suffice to summarize a fundamental principle. We're still at a point where some individual cases are understood, but we don't have a handle on any common mechanisms tying them together.
So what, exactly, does the hox system regulate and how? Again, understanding of this area has not yet reached the point at which a few sentences would suffice to summarize a fundamental principle. We're still at a point where some individual cases are understood, but we don't have a handle on any common mechanisms tying them together.


 Sadly, this forces us to get into the ParaHox genes, a task we had hoped to avoid. Our melancholy is due to: (a) our minimal understanding of ParaHox and (b) a depressing suspicion that the underlying theory is a gross and misleading oversimplification. As we lack any inclination to get creative here, we will fall back on the mythic language of reviews and textbooks. To us, this stuff sounds a bit like a Bronze Age creation myth:
Sadly, this forces us to get into the ParaHox genes, a task we had hoped to avoid. Our melancholy is due to: (a) our minimal understanding of ParaHox and (b) a depressing suspicion that the underlying theory is a gross and misleading oversimplification. As we lack any inclination to get creative here, we will fall back on the mythic language of reviews and textbooks. To us, this stuff sounds a bit like a Bronze Age creation myth: 
