Hox Genes - 1
A Really, Really Superficial Introduction to Evo-Devo
 The story of evo-devo is told, in very readable form, in Sean Carroll's 2005) recent book on the topic. Buy this book. If possible, do not bother finishing this essay. Just get the book. You don't even have to look it up: here's the link to the Amazon.com page. The rest of this discussion is mostly our clumsy attempt to explain what we learned there and elsewhere on this topic. Don't waste your time on it. Buy the book.
The story of evo-devo is told, in very readable form, in Sean Carroll's 2005) recent book on the topic. Buy this book. If possible, do not bother finishing this essay. Just get the book. You don't even have to look it up: here's the link to the Amazon.com page. The rest of this discussion is mostly our clumsy attempt to explain what we learned there and elsewhere on this topic. Don't waste your time on it. Buy the book.
Evolutionary developmental biology, "evo-devo," starts from the obvious: organisms get to be what they are through development as embryos, larvae, or whatever we label the stage(s) between fertilized egg and adult. Some embryos -- usually a minority -- will complete the process and become reproducing adults. However, almost nothing which happens after the embryo becomes an adult can have much effect on the biology of the organism. That's what "adulthood" means. Of course, natural selection continues throughout life; but what is selected are the characteristics acquired during development. Development is how we become what we are. It follows that, if we want to know how evolution makes changes, the proper focus is comparative embryology. If we want to get to the mechanics of these changes at a really fundamental level, then we must focus on comparing the way in which genes are regulated in the embryos of different organisms.
In fact, we can (as opposed to Kant) go a little further on pure reason. In animals, every cell has essentially the same complement of genes, and essentially all of those genes have some kind of homologue in every other animal. 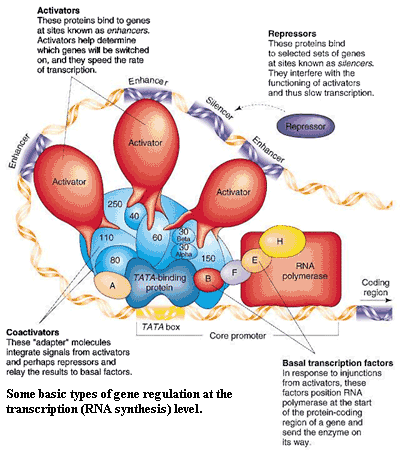 What separates us from chimpanzees can't be the miniscule differences in the sequences of the basic enzymes and structural proteins. The key is the regulation of those genes in the embryo. Carroll (2005a). One recent study could be read for the proposition that, even in protein-coding regions of DNA, the evolution of DNA sequence is primarily driven by the requirements for regulating when and where each gene is expressed (i.e. transcription and post-transcriptional processing in the nucleus), rather than by the biological function of the gene product. Parmley et al. (2007).
What separates us from chimpanzees can't be the miniscule differences in the sequences of the basic enzymes and structural proteins. The key is the regulation of those genes in the embryo. Carroll (2005a). One recent study could be read for the proposition that, even in protein-coding regions of DNA, the evolution of DNA sequence is primarily driven by the requirements for regulating when and where each gene is expressed (i.e. transcription and post-transcriptional processing in the nucleus), rather than by the biological function of the gene product. Parmley et al. (2007).
That's all off topic and rather too abstract. The point is that, even in the simplest organisms, genes don't just crank out RNA at random -- particularly in the embryo. Gene expression has to be carefully controlled. It must occur only at the right time during development and only in the right places -- just the right genes to make a leg, in the right sequence, in only the right places on the body, in the correct orientation, and hooked up correctly to the various nerves, muscles, blood vessels, and glands which integrate the leg with the rest of the adult organism. How is it done?
Evo-devo is still sketching the broadest outlines of the answer, but amazing progress has been made in the last decade. The best-understood part of the story -- arguably the only understood part of the story -- is anteroposterior axis "patterning" by hox genes. In other words, we're starting to get answers to the following fundamental question: how does an embryo tell its head from its ass? Of course, many adults never do seem to get this right. Fortunately, embryos rarely get involved in politics or religion, so they usually do a much better job than adults. Not only do embryos eschew these inherently inversionary activities, but they also have a special set of genes, the hox genes, which help them along.
In researching this section we ran into an unreasonably large proportion of exceptional papers -- too many to praise individually. This field has not only attracted some of the brightest and best, but some very engaging writers as well. We have marked a few of these papers with one or two asterisks when cited. Most can be found on the web. |
Hox genes are a subset of the homeobox genes. Some of you will doubtless attempt to claim that you've never heard of hox genes. We are not so gullible as to believe this. Yet it is possible -- just -- that you have temporarily mislaid this information, along with the birthdays of your parents and the name of your cat. We will mumble about them for a bit and perhaps you will recall what you have learned from some more reliable source.
Our approach uses three iterations. First, we offer some definitions and a few sentences of completely over-simplified noise -- the traditional high-school textbook stuff, without the (often very useful) diagrams. Next, we take a very brief look at some general biochemical facts about hox genes (You didn't think we were still talking about your cat, did you?). Then, having had about all the organization we can possibly stand, we will go back to doing things in the more customary Palaeos fashion. That is, we will take a long, meandering stroll through the metazoan cladogram, and point out some of the interesting sights along the way. Finally, if we have arrived at any conclusions in the process, we will probably state them. If not, we will end more or less abruptly: like this.
| Numerical |
Drosophila |
Abbreviation |
Notes |
| Hox1 |
labial |
lab |
Beginning of anterior class
and ANTP complex in Drosophila) |
| Hox2 |
proboscipedia |
pb |
|
| Hox3 |
(Zerknüllt, Bicoid) |
(zen, bcd) |
Hox3 class. Hox3 is probably its own
"upper middle class" homology group.
Drosophila has two hox3 homologues,
but neither is homeotic. |
| Hox4 |
Deformed |
Dfd |
Beginning of middle class |
| Hox5 |
Sex combs reduced |
Scr |
Insects have a "hox 5½,"
fushi
tarazu (ftz), which is not homeotic |
| Hox6 |
Antennapedia |
Antp |
Hox6-8 are homologues of Antp,Ubx
and AbdA as a group. That is, all 6
and ftz) derive from a single "lower
middle class" gene in Urbilateria. |
| Hox7 |
Ultrabithorax |
Ubx |
Beginning of UBX complex in
Drosophila |
| Hox8 |
Abdominal A |
AbdA |
|
| Hox9 |
Abdominal B |
AbdB |
Begining of posterior class |
| Hox10, 11 ... |
No arthropod homologues -- or (more
exactly) AbdB is homologous to all
posterior hox. |
Hox Genes: The hox literature is filled with bad nomenclature, starting with the term "hox" itself. A hox gene originally referred to any gene coding for a protein with a particular DNA-binding motif, the homeodomain. At the time, this was a small family, clearly identified with homeotic mutants of the fruit fly Drosophila. We'll explain the term homeotic later. Unfortunately, it turned out that the class of genes coding for homeodomain-containing proteins was vastly larger than the family of genes involved in homeotic mutations. As a result the term "hox" became utterly ambiguous. It could mean all genes which coded for homeodomain-containing proteins, or just the ones implicated in homeotic mutations. Watch out for this in the literature. We will use hox to mean the small set of genes involved in the homeotic mutations and their closest relatives [11] If we mean homeobox genes in general, we will say so. Recently, writers have introduced the term Hom to refer just to the homeotic (hox) genes. This terminology has not really caught on. We hope it will, but it hasn't. In any case, the numerical designations for hox genes are hox1, hox2, etc. For historical reasons, we're stuck with that system.
Notation: Historically, it was also customary to use the same name for a gene and the protein it coded. The gene was italicized. The protein was not. This sounds easy, but in fact the convention is cumbersome. Also, we are often sloppy and say things like "thoracic hox4 regulates limb differentiation." The truth is that, most of the time, we don't know whether or not hox4 is actually expressed in the thorax. Perhaps it is expressed elsewhere, and the hox4 mRNA or hox4 protein is transported to the thorax. We originally tried to stick to the old convention, because it promotes rigorous thinking; but it simply gets too awkward.
Fly names: When hox genes were first discovered in Drosophila, no one knew how, or even whether, the genes were related. Each hox gene was given a name by the discoverer of the corresponding mutant phenotype. Later, when it became clear what was going on, and hox-like regulation was investigated in other animals, things had become more organized. Hox genes were then given numbers. However, there are one hell of a lot of fruit fly workers out there, and the old names are widely used. Get used to them. In any case, as indicated in the notes to the table, the homology with vertebrate hox genes is not always exact. Thus, when speaking of lophotrochozoans in general, and arthropods in particular, it is actually more accurate to use the fly names.
Abbreviations: Also, the fly abbreviations all have initial capital letters except lab, pb, zen/bcd, and sometimes abdA. Use of abdA vs. AbdA is wildly inconsistent. We assume that this practice also has some obscure historical justification. Then again, it may be deliberate perversity. Fruitfly workers tend to enjoy that sort of thing. In any case, we've decided to jettison the capital letters along with italics unless we are giving the exact name of a particular gene in a particular organism (which we almost never do).
Homology classes: It is often convenient, and sometimes essential, to treat hox genes in groups. Conventionally, there are three or four homology classes: anterior, posterior, middle/central, and hox3. The reasons for this will become obvious as we go along. Molecular biologists tend to use the term orthology instead of homology because they don't understand what homology means. If you think we're simply being rude, see 14.2. Paralogy, Homology (Wikipedia), or Homology in Molecular Phylogenetics. The last one almost has it right. If you care, see the glossary entries. For practical purposes, paralogous genes are homologous genes in the same organism. Orthologous genes are homologous genes in different organisms. We tend to use homologous in both cases.
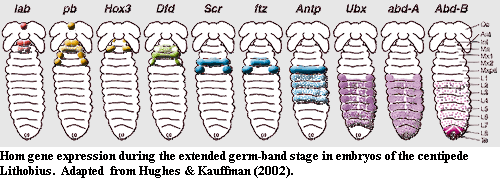 We'll postulate an ideal hox animal with none of the complications found in real animals -- call it Hoxazoon. Hoxazoon has a dozen or so hox genes. An ideal hox system has the following properties.
We'll postulate an ideal hox animal with none of the complications found in real animals -- call it Hoxazoon. Hoxazoon has a dozen or so hox genes. An ideal hox system has the following properties.
1) Genetic colinearity: All of the hox genes are found on one chromosome, one after the other, separated only by promoter/repressor regions. Hox1 is located on the 3' end of the cluster (i.e. "first"), while hox14 is on the 5' end ("last").
2) Anatomical colinearity: During development, the Hox genes are expressed in the same order along the anterior to posterior axis: hox1 toward the front of the head, hox14 at the tail end, as in the figure from Hughes & Kauffman 2002)*.
3)
Temporal colinearity: Generally, hox genes are also expressed in the same sequence in time -- hox1 first, hox14 last.
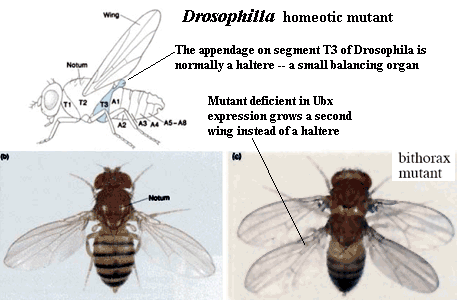 4) Evolutionary Conservation: Hox genes are quite highly conserved. In some cases, the mouse and fruit fly genes are practically identical. That's what makes it possible to use the same names for these genes in different animals without much loss of information.
4) Evolutionary Conservation: Hox genes are quite highly conserved. In some cases, the mouse and fruit fly genes are practically identical. That's what makes it possible to use the same names for these genes in different animals without much loss of information.
5) Homeotic transformation: You have doubtless been told by someone that the hox genes are responsible for "anteroposterior patterning" of the embryo. Although we used that language ourselves in the last section, it is actually a poor choice of words. What hox genes actually do is this: they allow embryonic development to progress using something close to an object-oriented programming language. Many high-level developmental signals are calls to an abstract class (e.g. a class of limbs). The object (the limb) is actually built using some subclass. The particular methods and data inherited by the subclass are specified by other inputs -- the developmental "context." This is where the hox genes come in. The particular combination of hox genes expressed in the target location specify what kind of object gets made: arm, leg, or even an intermediate form. The hox genes thus specify the particular methods and attributes included in the object.
As shown in the image, a homeotic mutant has the wrong pattern of hox expression and therefore gets the wrong kind of limb. Since JAVA programming may also be unfamiliar territory, we'll back away from the analogy; but it's a much more powerful powerful metaphore then "patterning" if you happen to have some experience in that area.
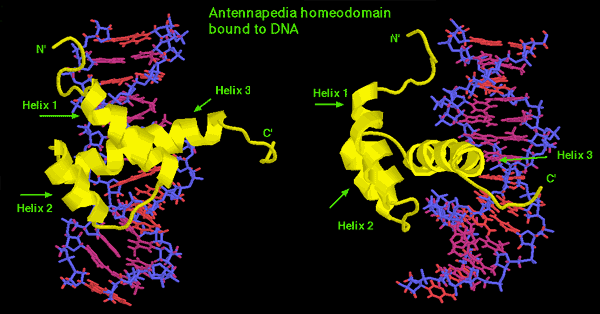 These five properties greatly simplify the machinery needed to develop an embryo. In essence, some master timing switch sends a message: "build me an appendage." That's all it needs to say. In a crustacean, for example, the cells receiving that same message will make a maxilliped, pedipalp, bristle, or whatever. The choice depends on which hox genes are active in the area. The message doesn't have to include detailed instructions about what kind of appendage to make or how to make it. The target cells respond to the message based on context -- the hox "code." This allows development -- and sometimes evolution -- to proceed in a simple, modular way.
These five properties greatly simplify the machinery needed to develop an embryo. In essence, some master timing switch sends a message: "build me an appendage." That's all it needs to say. In a crustacean, for example, the cells receiving that same message will make a maxilliped, pedipalp, bristle, or whatever. The choice depends on which hox genes are active in the area. The message doesn't have to include detailed instructions about what kind of appendage to make or how to make it. The target cells respond to the message based on context -- the hox "code." This allows development -- and sometimes evolution -- to proceed in a simple, modular way.
The general tells his adjutant, "get me a car." Depending on where they are and what unreasonable time frame the general has imposed, the adjutant decides what depot to call. He calls the sergeant at the depot and orders a suitable car. The sergeant decides what's available, and tells off a private to drive the car to the general. The private decides whether the car needs gas and how to get to the general. At each step, the instructions are simple and efficient, with execution dependent on context. The hox "code" provides that context. Unless something goes seriously wrong, the code ensures that the fruit fly grows a pair of halteres, rather than a second pair of wings. The general departs in a conveyance appropriate to his exalted rank (perhaps a Hom-V), rather than a water buffalo.
This idealized, linear version of hox regulation is not at all unique. Embryonic development, computer programming, and military structure are all methods for organizing incredibly complicated systems to acheive precise, predictable results in all kinds of unpredictable circumstances. The same basic elements are present in sophisticated versions of each: carefully graded decision-making heirarchies, modularity, error-checking and reporting, feedback loops, and some limited number of single-function specialist units.
CONTINUED ON NEXT PAGE
page ATW070513
checked ATW070725
 The story of evo-devo is told, in very readable form, in Sean Carroll's 2005) recent book on the topic. Buy this book. If possible, do not bother finishing this essay. Just get the book. You don't even have to look it up: here's the link to the Amazon.com page. The rest of this discussion is mostly our clumsy attempt to explain what we learned there and elsewhere on this topic. Don't waste your time on it. Buy the book.
The story of evo-devo is told, in very readable form, in Sean Carroll's 2005) recent book on the topic. Buy this book. If possible, do not bother finishing this essay. Just get the book. You don't even have to look it up: here's the link to the Amazon.com page. The rest of this discussion is mostly our clumsy attempt to explain what we learned there and elsewhere on this topic. Don't waste your time on it. Buy the book.

 4) Evolutionary Conservation: Hox genes are quite highly conserved. In some cases, the mouse and fruit fly genes are practically identical. That's what makes it possible to use the same names for these genes in different animals without much loss of information.
4) Evolutionary Conservation: Hox genes are quite highly conserved. In some cases, the mouse and fruit fly genes are practically identical. That's what makes it possible to use the same names for these genes in different animals without much loss of information. These five properties greatly simplify the machinery needed to develop an embryo. In essence, some master timing switch sends a message: "build me an appendage." That's all it needs to say. In a
These five properties greatly simplify the machinery needed to develop an embryo. In essence, some master timing switch sends a message: "build me an appendage." That's all it needs to say. In a 