
| Palaeos: |  |
Eukarya |
| EUKARYA | Eukarya Notes |
| Page Back | Unit Back | Unit Home | References | Glossary | Pieces |
| Page Next | Unit Next | Life | Dendrogram | Taxon Index | Time |
Life |--Eubacteria `--+--Archaea `--Eukarya |--Metamonada `--+--Discicristata `--+--Rhizaria `--+--+--+--Alveolata | | `--Chromista | `--Plantae `--Stem Metazoa |--Fungi `--Metazoa |
Eukarya General Introduction Lists Organization Origins of the Eukarya Introduction Cell Membranes and Walls Cytoskeleton General Metabolism Internal Membranes Chromosome and Genome Abrupt Concluding Remarks |
[1] Martin & Russell (2002) are particularly serious offenders. Two examples: Martin & Russell state: "Archaebacteria synthesize their isoprenoids via the condensation of IPP and its isomer, dimethylallyl diposphate, C5 units that are synthesized from acetyl-CoA via the MVA pathway (Langworthy et al. 1982). Notably, Eubacteria also synthesize isoprenoids, but they do so through a completely unrelated pathway, the DXP pathway, intermediates of which are precursors for thiamin diphosphate and pyridoxal diphosphate biosynthesis (Lange et al. 2000)." Actually Lange et al. (2000) (the "al." include Martin himself) note that the MVA pathway is nearly universal -- Eubacteria simply have the DXP pathway in addition. Second, Martin & Russell make the usual observation that Eubacteria have ester lipids while Archaea have ether lipids and that Eubacteria almost all have murein walls. They therefore conclude that "there is no similarity whatsoever in the components with which Archaebacteria and Eubacteria uniformly compartmentalize their cytosol from the environment ... ." Yet there exist bacteria, which we will discuss at length anon, who simultaneously (a) lack murein and (b) possess lipids containing both ester and ether linkages (in the same molecules). Fuerst (2004).
[2] L-forms have decreased dependence on FtsZ, the prototypical bacterial cell division signal. FtsZ is also the homologue of neomuran tubulin, so we will meet it again. For the moment, it's sufficient to note that the loss of the outer membrane frees up FtsZ to do other things. Similarly, the precursor of actin, MreB, normally forms a spiral cord under the outer membrane in Gram-negative bacteria. Gitai et al. (2004). Thus, MreB, too, must adapt to a different environment.
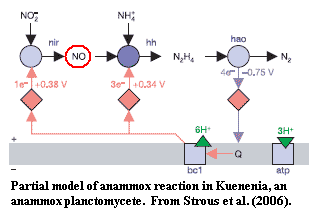 [3]
Sun & Liao's discovery of a connection between VDACs and NO-mediated cell
signaling is suggestive here because anammox planctomycetes, in particular,
generate NO as an intermediate in the anammox reaction.
Strous et al.
(2006). There seems to be some connection between the evolution of the
Neomura and weird nitrogen metabolism. We can't even speculate what that
connection might be, but we will see it repeatedly in this discussion.
[3]
Sun & Liao's discovery of a connection between VDACs and NO-mediated cell
signaling is suggestive here because anammox planctomycetes, in particular,
generate NO as an intermediate in the anammox reaction.
Strous et al.
(2006). There seems to be some connection between the evolution of the
Neomura and weird nitrogen metabolism. We can't even speculate what that
connection might be, but we will see it repeatedly in this discussion.
[4] Archaea seem to lack Lipid A-associated genes. However, Archaea, in general, have a very small repertoire of lipid synthesizing genes. Karlin et al. (2005).
[5] To be perfectly honest, we have run into this statement, a number of times, in places like Wikipedia; but we have not been able to confirm it. What we have confirmed is that the cell wall proteins are rich in proline and glutamate and resistant to disruption by detergents, all arguing that the protein is polar and tightly linked by hydrogen bonds. Some, but not all, planctomycete cell "walls" are also bound together by cysteine disulfide bridges. Fuerst (1995). All of this sounds like the kind of environment which would be consistent with glycoprotein, but we can't promise that the statement is accurate.
[6] The term "dehydrogenase" implies an oxidation reaction. It is important to remember that the reaction can proceed either way, depending on the thermodynamics of the reaction, the concentration of substrate, and the coupled reaction of the cofactor. In our case, the biologically relevant direction is reduction of the ketone (C=O) to an alcohol (C-OH). This reduction is thermodynamically disfavored, but is driven by coupling the reduction with the highly favored oxidation of the nucleotide cofactor, e.g. NADH+ to NAD. Thus, the enzyme is constructed in a very simple way: the dehydrogenase holds the ketone. The Rossman fold holds the nucleotide. The thermodynamics of the protein's conformation, usually assisted by a divalent ion of zinc or magnesium, favor a close approach of the two, and the deed is done.
[7] To be frank, we know of another problem area, also involving lipids. This relates to the pathway by which isoprenoids are synthesized (deoxyxylulose vs. mevalonate pathways). We're going to skip this one, inter alia, because it deals more with Archaea than Eukarya and because the details of the deoxyxylulose pathway are not fully understood. See Bonanno et al. (2001) for a slightly out-of-date review.
[8] Thus becoming a purine polar para proton-plucking partial protein. By an interesting coincidence, GDH also happens to work in a pathway next door to the pathways of G1PDH and G3PDH. GDH catalyzes the oxidation of glycerol to dihydroxyacetone. If you recall, dihydroxyacetone phosphate (DHAP) is the substrate for G1PDH and G3PDH. This being a mere footnote, we might wildly speculate that this has very deep, pre-LUCA, phylogenetic significance. We won't, because it would take us on a long biochemical tangent. But think of the implications of having (originally) a single enzyme, close to the base of about 40% of life's most critical metabolic pathways, which can act on almost any C-O bond, and whose action (oxidation or reduction) intrinsically depends on (a) whether the adjacent hydroxyl is phosphorylated (b) the availability of divalent cations in the medium and (c) the oxidation state of a general-purpose electron donor/acceptor. Given just that one enzyme and a kinase, one could reconstruct the core of a very plausible pre-LUCA biochemistry.
[9] Interestingly, it seems that Rhodopirellula has a particularly broad selection of dehydrogenases, over which it exerts fine metabolic control. Gade et al. (2003). This is what one might expect of these accomplished oligotrophs, who are able to live almost anywhere, on almost any substrate.
[10] "Both reactions involve a deprotonation of substrate, followed by hydride transfer to NAD+. Both enzymes use a Zn2+-activated water molecule with structurally aligning ligands to carry out the deprotonation step." Bartlett et al. (2003).
[11] Certain ectosymbionts of Euplotidium, apparently also verrucomicrobes, also may contain tubulin. Petroni et al. (2000). In addition, Cho et al. (2004) have recently reported a new group of baritones, the Lentisphaerae, tucked in between the Verrucomicrobia and the Chlamydiae, which have similar, if less dramatic, structures. Like eukaryotic tubulins, the baritone Atub comes in two sorts, Atub a and Atub b. This allows it to form heterodimers similar to the α/β dimers of eukaryotic tubulin. However, neither one of the Atub monomers is more closely related to one particular eukaryotic tubulin. Michie & Löwe (2006). The heterodimer system has a number of regulatory advantages. So, it is not surprising that Prosthecobacter and eukaryotes should arrive at similar solutions to the same problem.
[12] A third possibility might be a bacterium with two copies of FtsZ, in which case one might get both FtsZ and tubulin. This is conceivable, but problematic, since FtsZ is associated with the origin of DNA replication in most bacteria. So long as FtsZ is performing that role, an aberrant FtsZ orthologue is likely to be fatal long before it becomes useful for something else. Thus, FtsZ-dependence for division must be reduced before we can talk sensibly about exaptation.
[13] MreB is abundant in Archaea. As far as we can tell, ParM is unknown in the Archaea. Accordingly, ParM may be a red herring for phylogenetic purposes.
[14] In particular, we assume (a) that anammox bacteria can fix ammonia for amino acids and (b) that Strous et al. (2006) are using the same gene abbreviations as Cabello et al. (2004). The Strous paper is the usual, hyper-compressed Nature "letter." All the hard data are hidden away in some inaccessible corner of the Nature site, and shuffled around periodically to keep anyone from finding it.
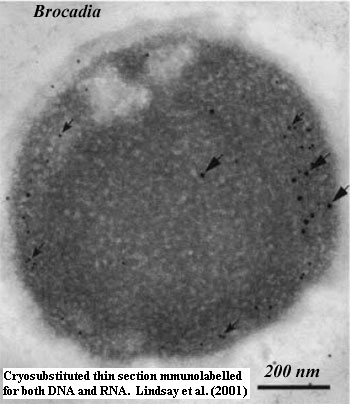 [15]
Lindsay et al. (2001) contains one of the most utterly intimidating
"Materials and Methods" sections we have ever seen. Here's but a small
sample:
[15]
Lindsay et al. (2001) contains one of the most utterly intimidating
"Materials and Methods" sections we have ever seen. Here's but a small
sample:
Thin sections of cryosubstituted Pi. marina, I. pallida, G. obscuriglobus and “Candidatus Brocadia anammoxidans” on nickel grids were floated onto a drop of PBS pH 7.5/0.2% fish skin gelatin/ 0.2% bovine serum albumin/glycine (0.02 M) (PBS/FBG) on a sheet of Parafilm for 5 min. They were then floated onto drops of primary antibody, mouse IgM anti-double-stranded/single-stranded DNA (Boehringer-Mannheim), diluted 1:20 in PBS/FBG for 1 h, washed on four drops of PBS/FBG and then placed on a drop of goat anti-mouse IgG+IgM coupled to 15 nm colloidal gold antibody (British Biocell International) diluted 1:20 in PBS/FBG for 1 h. Grids were then subjected to 4× PBS washes of 4 min each and 4×2-min water washes. The grids were dried and sections stained as described with methanolic uranyl acetate and Reynolds lead citrate.
Translation for the uninitiated: carefully replace all water molecules of flash-frozen cells with ultrapure organic solvent, embed cells in a resin and cut invisibly thin sections, mount thin sections on tiny grids, treat for precise intervals on individual drops of two different antibody solutions (a triple-antibody procedure!), multiple washes, dry carefully, double stain with incredibly toxic metal solutions -- all without warping, disturbing, contaminating, or otherwise screwing up the invisible specimen on its tiny grid. In many cases the procedure was even more complex, involving an additional set of antibody labels for RNA. After a little web sleuthing, we concluded that Richard I. Webb, of the University of Queensland Centre For Microscopy And Microanalysis, was probably the one who originally worked out the mechanics of doing this kind of thing. We've seen some of his other work and are thoroughly impressed.
[16] Maybe the wrong membrane. There have been suggestions that the anammoxosome membrane is homologous to the nuclear membrane of Gemmata. We're not sure where this comes from, but it makes sense. The anammox reaction requires a complex membrane, and the organelle does contain low levels of DNA (see the image of Brocadia at [15]). Lindsay et al. (2001).
[17] Actually dodecameric. The complex consists of a protease and separate ATP-binding chaperone unit, rather than having both in one molecule. As in the 20S proteasome, two of these 12-mers face each other to form a barrel with two ends. Azim et al. (2005). This separation of chaperone and protease jobs in the HslV multimer may be closer to the way in which archaeal and eukaryotic proteasomes actually function than is the operation of the actinobacterial 20S unit.
[18] We have to weasel on this, just a bit. The group includes at least one member of the Acidobacteria, whose phylogenetic position is rather vague. In the past few years, they have been assigned to the basses, baritones, tenors, and even low altos (low G+C Gram positives) -- in fact everywhere except the upper altos where the Actinobacteria are found.
[19] In particular, we looked to see whether something similar occurs in the actinobacterium Mycobacterium, since it also has a great deal of DNA in a small space. The chromatin of Mycobacterium appears condensed in conventional preparations, but is well-dispersed using cryosubstitution methods. Paul & Beveridge (1992). Planctomycete chromatin is condensed even in cryosubstituted preparations. Lindsay et al. (2001). Another possible case lies at the opposite end of the DNA concentration scale, in the behemoth bacterium Epulopiscium. However the details of that case are poorly known, as are the phylogenetic affinities of this very odd bug. Bressler & Fishelson (2004); Angert (2005).
| Page Back | Page Top | Unit Home | Page Next |