
| Palaeos: |  |
Eukarya |
| EUKARYA | Eukarya Origins - 6 |
| Page Back | Unit Back | Unit Home | References | Glossary | Pieces |
| Page Next | Unit Next | Life | Dendrogram | Taxon Index | Time |
Life |--Eubacteria `--+--Archaea `--Eukarya |--Metamonada `--+--Discicristata `--+--Rhizaria `--+--+--+--Alveolata | | `--Chromista | `--Plantae `--Stem Metazoa |--Fungi `--Metazoa |
Eukarya General Introduction Lists Organization Origins of the Eukarya Introduction Cell Membranes and Walls Cytoskeleton General Metabolism Internal Membranes Chromosome and Genome Abrupt Concluding Remarks |
 Now
the point of all this, in case you wondered, is that the evolution of the
nucleus and of chromatin can be understood in this simple way. We may find
a few bacteria with precursors of some of the refinements which neomuran
chromatin made possible. However, all that sort of thing is secondary.
The real issue is "What are they doing about the glop?" With the issue so
elegantly reformulated, the evidence points unerringly in the direction of the planctomycetes.
Regardless of whether the Gemmata nuclear body membrane is a "genuine"
nucleus, it contains all of the cell's DNA and shields the rest of the cell from
the effects of having a large polyelectrolyte in a small space.
Now
the point of all this, in case you wondered, is that the evolution of the
nucleus and of chromatin can be understood in this simple way. We may find
a few bacteria with precursors of some of the refinements which neomuran
chromatin made possible. However, all that sort of thing is secondary.
The real issue is "What are they doing about the glop?" With the issue so
elegantly reformulated, the evidence points unerringly in the direction of the planctomycetes.
Regardless of whether the Gemmata nuclear body membrane is a "genuine"
nucleus, it contains all of the cell's DNA and shields the rest of the cell from
the effects of having a large polyelectrolyte in a small space.
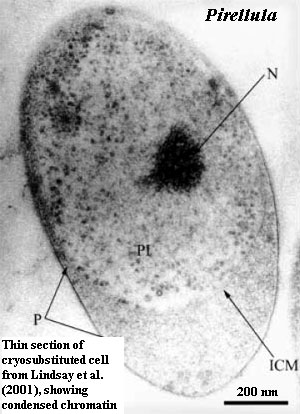
Similarly, regardless of whether the planctomycetes have "genuine" histones, they have condensed chromatin. This is an unusual feature in bacteria, and is generally found, if at all, in specialized structures which are not transcribing RNA. So far as we have been able to determine, condensed chromatin occurs in transcriptionally active eubacterial cells only in Caulobacter swarmers (Bendich & Drlica, 2000) -- and in all planctomycetes (except possibly Isosphaera). Fuerst (2005); Lindsay et al. (2001) . [19]
Notice that the planctomycete solution to having a lot of DNA in a small space is to compact most of the DNA into an even smaller space. This is the eukaryote solution to the glop problem, and the planctomycetes seem to be the only non-neomuran group which has adopted this approach. Since all, or perhaps almost all, planctomycetes use the same strategy, we are probably safe in assuming that they and their ancestors have been doing this for over two billion years. So, this is not mere scenario-building. By several orders of magnitude, the baritones are the most likely group to have evolved a strain with neomuran-style histones and nucleosomes at some point in their unending struggle with the fundamental chemistry of DNA.
The guts and feathers of modern biology is all in transcriptional and post-transcriptional control, so you will undoubtedly be awaiting some extended discussion of this subject. If so, forget it. Most of the really interesting stuff seems to have come out in just the last two or three years. Perhaps more to the point, we really haven't read enough of it, even by our own, notoriously relaxed standards of scholarship.
The general trend here seems to be consistent with the rest of the newer evidence. That is, the yawning evolutionary abyss between Eubacteria and eukaryotes is looking a little less intimidating each month. Like the Grand Canyon, it is unquestionably a big ditch; but a careful study of the details will locate any number of ways across, so long as you watch your footing and allow enough time. The problem now is figuring out which path life actually took. What we can be increasingly certain of is that life did not abruptly sprout wings, use a magic levitating spell, or otherwise cheat on the fundamental rules of evolution. Even in the area of transcriptional control, a pattern of evolutionary connections between the three domains of life is beginning to emerge. See, for example, Hickey et al. (2002) (chaperones), Hand et al. (2005) (secretory proteins), Thaw et al. (2006) (amino acid regulators).
The trick, as so often in the history of evolutionary thinking, is to avoid jumping to conclusions. Think of the evolution of birds, before feathered dinosaurs were discovered; tetrapods, before Ichthyostega; or humans, before Australopithecus. In each case, prior to the key fossil discoveries, the evidence seemed to indicate some sharp evolutionary discontinuity, a "quantum" transition (Cavalier-Smith makes precisely this tired analogy). However, in each case, the actual course of evolution turned out to be the usual plodding, step-wise affair, with all kinds of bushiness in the tree. We suspect the origin of eukaryotes was no different -- except that, as we said at the beginning -- two billion years is a hell of a long time.
And, on the subject of not leaping to conclusions, are we serious about the evolutionary importance of the Planctomycetes? Or has this all been an extended send-up of Cavalier-Smith? That's a good question. We still don't know the answer and have vacillated a good deal over the course of writing this piece. For what it may be worth, we are completely convinced by Cavalier-Smith's main point, that the Eubacteria are paraphyletic. On the other hand, we have exaggerated our aversion to horizontal gene transfer, just to see where that path might lead. It truly does seem to point to the baritones.
 However,
there are ... problems. For one thing, gene transfer from plastids to nuclear
genomes is probably too well established to dismiss quite as easily as our
rhetoric might suggest. For another, the Planctomycetes seem sometimes to
make good proto-eukaryotes and sometimes good proto-archaeans. What they
don't seem to make are good proto-neomurans. For example, their internal
membrane system places them very close to an extrapolated line drawn between
Eubacteria and a Eukarya. By contrast, planctomycete C1
and nitrogen
metabolism seems to place them mid-way on a line between Proteobacteria and
Archaea. Unfortunately, Archaea don't have nuclei and eukaryotes don't have these
sorts of metabolic systems. This evolutionary dissonance makes us anxious and fretful. But,
as we said only one paragraph ago, it's still too soon.
However,
there are ... problems. For one thing, gene transfer from plastids to nuclear
genomes is probably too well established to dismiss quite as easily as our
rhetoric might suggest. For another, the Planctomycetes seem sometimes to
make good proto-eukaryotes and sometimes good proto-archaeans. What they
don't seem to make are good proto-neomurans. For example, their internal
membrane system places them very close to an extrapolated line drawn between
Eubacteria and a Eukarya. By contrast, planctomycete C1
and nitrogen
metabolism seems to place them mid-way on a line between Proteobacteria and
Archaea. Unfortunately, Archaea don't have nuclei and eukaryotes don't have these
sorts of metabolic systems. This evolutionary dissonance makes us anxious and fretful. But,
as we said only one paragraph ago, it's still too soon.
While the state of our substantive knowledge is still frustratingly incomplete, this long exercise has improved our confidence in two methodological assumptions mentioned at the outset. First, we tried to stick to actual facts about actual organisms, rather than base our analysis on a priori estimation of the likelihood that things would happen in a particular order. We weren't completely faithful to this commitment; but, by and large, it turned out well.
Second, the strategy of giving structure priority over sequence was even more productive than we had anticipated. True, this is a gong we have been beating loudly for years. But our faith was beginning to waiver. Sequence data have been growing explosively in the last five years, and the tools for handling sequences are becoming more sophisticated daily. Many of those tools are, quite frankly, well beyond our mathematical experience; and we felt like a Neanderthal trying to critique the engineering of a radio. Fortunately, resources such as PDB are also growing at a fast clip, and we can hope to have structural information comparable to our present sequence data in less than a decade. In the meanwhile we can amuse ourselves reading the growing number of papers which apologetically jettison completely useless sequence analyses because the structural comparisons make the correct answers compelling and obvious.
| Page Back | Page Top | Unit Home | Page Next |