|
|
Cladistics |
| Systematics |
Phylogenetic Systematics |
Cladistics: Phylogenetic Systematics
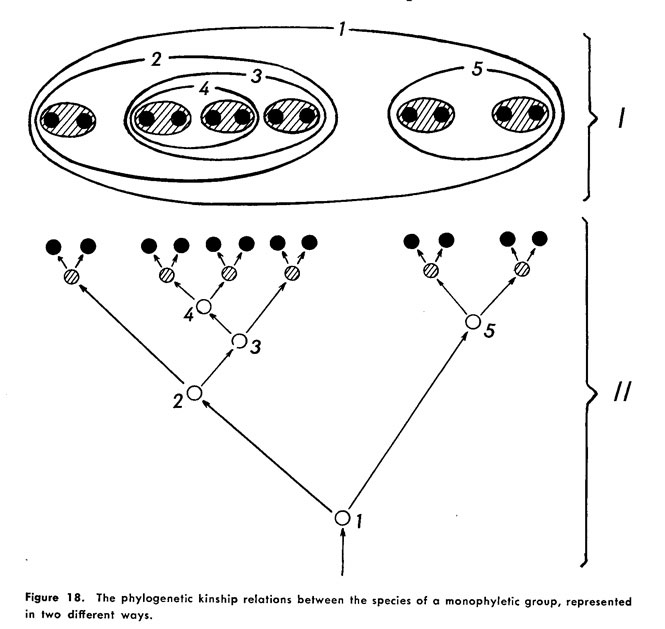
Hennig's two types of cladistic diagrams, from Phylogenetic Systematics, figure 18, showing nested clades. The caption reads: "The phylogenetic kinship relationship between the species of a monophyletic group, represented in two different ways." The top drawing (I) shows a set-based, nesting diagram, while the bottom (II) shows a branching tree diagram. This diagram shows the interconnection between branching diagrams and nesting diagrams, but remains equivocal about which is best for displaying the evolutionary relationships phylogenetic methods aim to uncover. - from Rebecca Shapley, Visualizing the Tree of Life |
History
"Phylogenetic Systematics" is a rigorous methodology first developed by German entomologist Willi Hennig in the 50s and 60s as a means of evaluating and reconstructing phylogenies if fossils are lacking (insects have a notoriously poor fossil record). Hennig's work was translated into English as Phylogenetic Systematics (University of Illinois Press, 1966), which remains a foundational text for modern phylogenetic studies. Nevertheless his work remained little known until the late 70s, when it was rediscovered and taken up by newer workers in the field such as James S. Farris, Walter Fitchand, and Herb Wagner, and applied in American and British paleontology. Note that Hennig never used the word "cladistics", which was coined by Mayr for an adherent of Hennig's school.
For a short while there was a rival school or schools, known as Pattern or Transformed Cladistics, which rejected Hennig's emphasis on phylogeny in favour of cladograms as tests of a phylogenetic reconstruction, rather than as anything to do with actual evolutionary history, and which had elements in common with phenetics. But such an approach lacked the appeal of Phylogenetic Systematics, and although the idea of hypothesis testing was retained in later forms of cladistics, the rejection of phylogeny was not.
Here we use Phylogenetic Systematics to refer to traditional, Hennigian cladistics and its later and current applications, as opposed to Pattern and Transformed Cladistics, Computational cladistics, and Phylogenetics.
Methodology
Phylogenetic Systematics focuses on identifying unique (derived) shared characteristics, called, synapomorphies. These are distinguished from primitive shared characteristics, which are called plesiomorphies. Only synapomorphies can provide information about the evolutionary history of a group; its phylogeny. The aim is to identify what Hennig called monophyletic groups, that is groups consisting of common ancestor and all its desendents. These are known as clades. This is represented in the form of the now familiar and ubiquitous tree like diagram called a cladogram. In contrast to evolutionary systematics, Phylogenetic Systematics only acknowledges monophyletic groups. The relation between the different clades is shown by a tree-like diagram or cladogram. Because there are any number of possible alternative cladograms (or evolutionary histories), only the simplest one, requiring the least number of changes, is chosen.
In short, Linnaean taxonomy and Evolutionary Systematics are concerned with both plesiomorphies and synapomorphies, whereas Phylogenetic Systematics is only concerned with synapomorphies only; Evolutionary Systematics is concerned with both monophyletic (also called holophyletic) and paraphyletic groups (made up of a common ancestor and only some of its desendents) , whereas Phylogenetic Systematics only with monophyletic groups. And unlike Evolutionary Systematics it does not use a specific Linnaean hierarchy of ranks (e.g. class, phylum). This is why cladists woin't even recognise Class Reptilia as a natural group; because reptiles evolved into two other taxa, birds and mammals, they are paraphyletic rather than monophyletic. MAK130320
In this context, the following definitions are used
Outgroup We have seen above that a clade is a branch on a tree of descent. Anything occurring outside that branch, further towards the root of the tree, is an outgroup.
The distinction is more than simply contextual: For example, in order to calculate the similarity of genome sequences, it is essential to include within the study one or, preferably, several taxa that lie outside the group in which we are trying to detect relationships. If we are interested in determining the relationships of tigers, we would use close relatives of tigers as our outgroups.
Outgroup comparison is the way we determine how widespread a particular feature may be, whether it is found only within the group (apomorphies) of interest, or beyond that group.
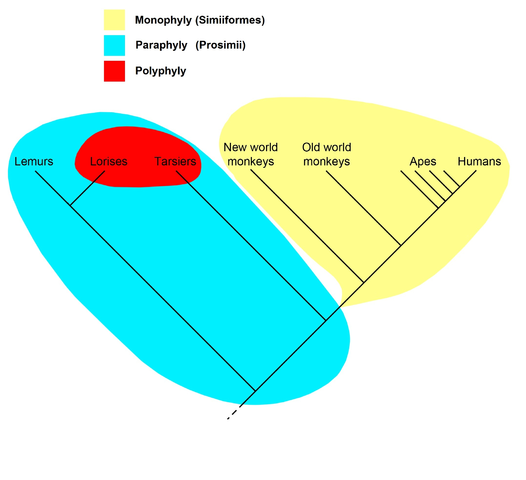
A cladogram of the primates with groups showing different phylogenetic units: Monophyly (Simiiformes), paraphyly (Prosimii) and polyphyly (the night active primates, the lorisis and the tarsiers). Diagram by Petter Bøckman, revised by Peter Brown, Creative Commons Attribution, Wikimedia |
Monophyly (Monophyletic Group) A monophyletic group is one which includes an ancestral species and all its descendants. It is a complete clade.
As we have seen, a monophyletic group can be extremely large and inclusive – for example, most people today would agree the legions of different kinds of insects comprise a monophyletic group – or quite small and exclusive – for example, the enigmatic sea spiders (class Pycnogonida).
Paraphyly (Paraphyletic Group) - A paraphyletic group is a clade lacking some of the descendant species.
Today there is a movement away from applying formal names to groups which are known to be paraphyletic, although some of the old taxa are still very useful even though they are now believed to be paraphyletic.
Perhaps the best example is the reptiles. Because both mammals and birds evolved from reptilian ancestors, but are not included in the class Reptilia, the latter is clearly paraphyletic and a cladistic purist might prefer not to use the name. However, the meaning and scope of the reptile class is still a very well understood and useful concept.
What is more, if we were to blindly enforce “Russian doll nomenclature” in this fashion, it seems unlikely the existing hierarchy of taxonomic ranks will cope.
Polyphyly (Polyphyletic Group) - A polyphyletic taxon is an “unnatural” assemblage of two or more clades, united by some characteristic which is not a primitive feature (plesiomorphy).
Groupings which are thought to be polyphyletic truly are avoided by taxonomists, which is one reason there are not too many familiar real examples. An artificial example is “warm blooded animals,” a group which includes both mammals and birds. However, both these groups arose, at different times, from cold-blooded (reptilian) stock: their warmbloodedness is not an apomorphy, but it evolved separately, and is different in detail.
Again, however, all is not plain and simple. Some taxa, even those with a long history of study such as the arthropods, are still subject to on-going controversy. Although most researchers are of the view that the Arthropoda are a “good” monophyletic clade, there remain a few who argue that the arthropod characteristics were arrived at separately by more than one lineage, and thus the group is polyphyletic. They are in a small minority, but some small doubt remains.
Chris Clowes 030219
From Wikipedia
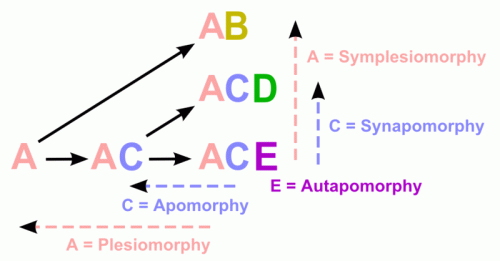
A plesiomorphy ("close form") or ancestral state is a character state that a taxon has retained from its ancestors. When two or more taxa that are not nested within each other share a plesiomorphy, it is a symplesiomorphy (from syn-, "together") of theirs. Symplesiomorphies do not mean that the taxa that have them are necessarily closely related. For example, Reptilia is traditionally characterized by (among other things) being cold-blooded (i.e. not maintaining a constant high body temperature), whereas birds are warm-blooded. Since cold-bloodedness is a plesiomorphy, inherited from the common ancestor of traditional reptiles and birds, and thus a symplesiomorphy of turtles, snakes and crocodiles (among others), it does not mean that turtles, snakes and crocodiles form a clade that excludes the birds.
An apomorphy ("separate form") or derived state is an innovation. It can thus be used to diagnose a clade - or even to define a clade name in phylogenetic nomenclature. One clade may have autapomorphies (from auto-, "self"), two sister-groups may have synapomorphies (from syn-, "together"). For example, the possession of digits that are homologous with those of Homo sapiens is an apomorphy within the vertebrates. The tetrapods can be singled out as consisting of the first vertebrate with such digits together with all descendants of this vertebrate (an apomorphy-based phylogenetic definition). Importantly, snakes and other tetrapods that do not have digits are nonetheless tetrapods: they descend from ancestors that possessed them.
A character state is homoplastic or "a homoplasy" if it is shared by two or more organisms but was not present in their common ancestor. It has evolved by convergence or reversion. Both mammals and birds are able to maintain a high constant body temperature (i.e. they are 'warm-blooded'). However, the ancestors of each group did not share this character, so it must have evolved independently. Warm-bloodedness is separately an apomorphy of mammals and birds, but it is not a synapomorphy of these two clades.
The terms plesiomorphy and apomorphy are relative; their application depends on the position of a group within a tree. An (aut)apomorphy of one clade is a plesiomorphy of each of its members.
- Wikipedia
An actual example of cladistic analysis, showing the way characters can be used to determine the phylogenetic relationship between taxa, is shown below:
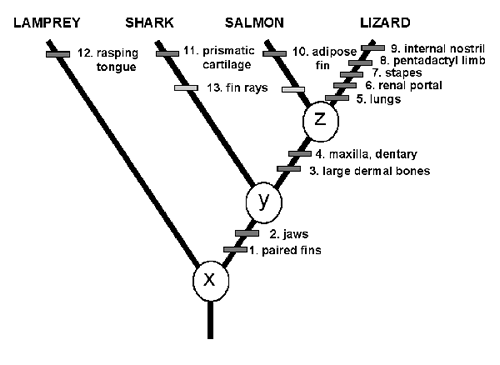
An example of a cladogram showing characters by which taxa are recognised. Characters 1 - 4 are synapomorphies, 5 - 12 are autapomorphies and 13 is an attribute seen in the salmon and the shark. Characters numbered 3 and 4 are synapomorphies suggesting that the lizard and the salmon shared a unique common ancestor 'Z'. It suggests that characters 3 and 4 arose in ancestor 'Z' and were inherited by the salmon and the lizard. Shared primitive characters (symplesiomorphies) are characters inherited from a more remote ancestry and are irrelevant to the problem of relationship of the lizard and the salmon. For example, the shared possession of characters 1 and 2 in the salmon and lizard would not imply that they shared a unique common ancestor because these attributes are also found in the shark. Characters 1 and 2 may be useful at a more inclusive hierarchical level to suggest common ancestry at 'Y'. With respect to the three-taxon problem (shark, salmon and lizard) then characters 1 and 2 are symplesiomorphies and they suggest nothing other than that the shark, salmon and lizard are a group. Similarly, characters 5 - 9 and 10 - 12 are autapomorphies and irrelevant to discovering relationships since they are each found in only one of the taxa. Sister-groups are discovered by identifying shared derived apomorphic characters (synapomorphies) inferred to have originated in the latest common ancestor and shared by descendants. These synapomorphies can be thought of as evolutionary homologies: that is, as structures inherited from the immediate common ancestor. Diagram and caption from Cladistics for Palaeontologists © Courtesy The Palaeontological Association. |
From Phylogenetic Systematics to Computational cladistics
In the decade or two from the mid-1980s onwards, this methodology was used with great effectiveness to work out the relationship between various taxa of living and fossil vertebrates, such as for example the descent of birds from dinosaurs, or the phylogenetic relationship between early reptiles (amniotes). This type of phylogenetic methopdology became the "gold standard" for determining evolutionary relationships.
But with the development of cheap computing, simple trees constructed from easily recognisable and well studied synapomorphies such as those described in the diagram on the right, have been replaced by a new methodology called computational phylogeny. This relies on statistical analyses of huge data matrixes, featuring hundreds of character states and throwing up literally millions of possible cladograms. It still uses all of the same principles as classical Phylogenetic Systematics, such as distinguishing synapomorphies from plesiomorphies, and homologies from homoplasies (convergences), in order to identify monophyletic clades, and selecting the best phylogenetic hypothesis on this basis, usually (although not always) by means of parsimony. But it often throws up totally different cladograms to those arrived at using the single tree method.
In recent years morphology-based computational cladistics has become assimilated into molecular phylogeny, the two together becoming the statistical science of phylogentics, in which parsimony and morphology play second fiddle to molecular sequencing. MAK130321



