
| Palaeos |  |
Glossary |
| Metazoa | Glossary H-N |
| Page Back | Unit Back | Unit Home | References | Glossary | Pieces |
| Page Next | Unit Next | Life | Dendrogram | Taxon Index | Time |
LIFE |--Eubacteria `--+--Archaea `--Eukarya |--Chlorobionta `--+--Fungi `--METAZOA |--Choanoflagellata `--Porifera |--Radiata `--+--Cnidaria `--Bilateria |--Deuterostomia `--Protostomia |--Ecdysozoa `--Spiralia |
The eventual aim is to combine all the glossaries distributed throughout the metazoan section. This will takes many years. |
A B C D E F G H I J K L M N O P Q R S T U V W X Y Z
helix-turn-helix: another diverse and important DNA-binding protein motif, including many transcription factors, not to be confused with helix-loop-helix. For figure and brief discussion see Helix-Turn-Helix.
hexactine: (sponge anatomy) a spicule with six rays. See Spicule Terms.
histogenesis: (embryology) formation of tissues.
holoblastic: (embryology) type of cleavage of the very early embryo in which the planes of cleavage divide the entire embryo.
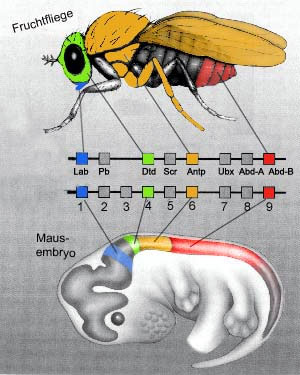 homeobox:
(molecular biology)
The DNA sequences corresponding to the homeodomain.
homeobox:
(molecular biology)
The DNA sequences corresponding to the homeodomain.
homeodomain: a particular DNA-binding protein motif containing about 60 amino acids. Proteins with a homeodomain are encoded by homeobox genes. The homeodomain proteins are important transcriptional regulators, particularly in embryonic development. They include the hox genes.
homeotic mutation: a mutation in which anterior structures are shifted to, or (more often) repeated in, more posterior parts of the body.
hom gene: alternative name for hox genes.
homology: two genes are homologous if they derive from a single ancestral gene. This isn't a precise term, because genes can pick up bits and pieces of other DNA through various types of transposition or errors during mitotic recombination. At some point -- a point which has no objective definition -- it is no longer homologous. Homology between structures is a concept in flux. The theoretical definition has always been essentially the same as homology applied to genes. That is, homologous structures are the phenotype resulting from homologous genes. However, for both historical and practical reasons, the definition is often confused with the operational tests of homology; "similarity," "congruence," and (sometimes) "conjunction." See, e.g. Witmer (1995); Hutchinson (2001). However the ultimate issue in homology is always ancestry. See also paralogy, orthology.
homonomous: anatomy. Having repeated, identical structures (usually said of body segments).
homoplasy: "convergent evolution" Similar structures/genes which are not homologous are homoplastic.
hox: see Hox genes for an extended discussion. Hox genes are homeobox genes with a special role in development. Homeobox genes are frequently found in clustered groups in the organism's DNA, always (almost) in the same order. The jaw-dropping and eyebrow-raising feature of hox genes is that their order along the organism's DNA corresponds to the physical order in which these genes are first expressed along the anteroposterior axis of the body during development. For example, the first member of the series is expressed in the anterior part of the head, and the last is expressed in the most posterior part (typically, the telson, or "tail"). The metazoans include animals with almost every imaginable variation on this theme, but arthropods, and vertebrates, and various other phyla all have essentially identical homeobox genes, in the same order, all expressed in (almost) the same order along the anteroposterior axis. It is almost universally believed that these homeobox genes specify the type of segment which will develop and the type of appendages (if any) that the segment will produce. The term hox is sometimes used to refer to all homeobox genes. However, hox is not the only class of homeobox genes. There are many others. In addition, some of the hox series are recycled later in development for other purposes, such as helping to specify the proximal to distal axis of the appendages.
hsp70, (biochemistry) a class of "chaperone" proteins which assist in the folding of other newly synthesized proteins. Hsp70 proteins have a particular target preference for hydrophobic amino acid sequences.
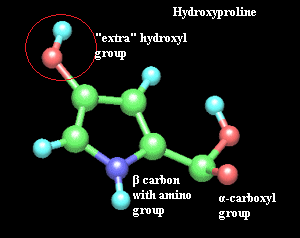 hydroxyproline:
(biochemistry) an atypical amino acid. Hydroxyproline is not found in the
genetic code. Instead it is made from proline enzymatically, after this
amino acid has already been incorporated into a protein. Hydroxyproline is
common in collagens.
hydroxyproline:
(biochemistry) an atypical amino acid. Hydroxyproline is not found in the
genetic code. Instead it is made from proline enzymatically, after this
amino acid has already been incorporated into a protein. Hydroxyproline is
common in collagens.
hypomorph: a mutant phenotype which is characterized by quantitative changes.
hypostome: "The hypostome is a ventral sclerotic plate between the antennulae and the antennae (with sclerotic extensions, named wings, which serve as holdfast for limb musculature), while the labrum is a soft, bulged outgrowth at the posterior end of the hypostome, only present in labrophoran Crustacea (most likely originating from the distal part of the mouth membrane, well developed in Agnostus pisiformis [a trilobite] ... . Accordingly, both structures are not the same and cannot be synonymized, thus." Stein et al. (2005).
instar: any discreet stage in a developmental series separated by successive molts, metamorphoses, etc.
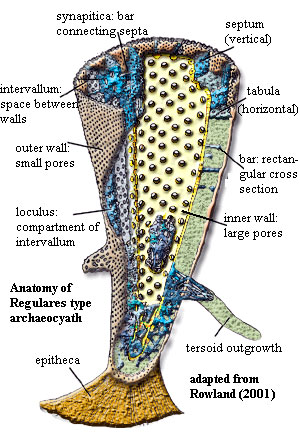 intervallum:
in archaeocyath anatomy, the space between the inner and outer walls.
intervallum:
in archaeocyath anatomy, the space between the inner and outer walls.
intron: part of a gene which is flanked on both sides by sequences coding for the corresponding protein, but which is not itself part of the coding sequence.
-ite: (Crustacea) a suffix applied to appendage elements, perhaps originally meant to suggest that the element was a secondary outgrowth from the main axis, but now essentially meaningless.
lab: abbreviation for labial, the hox1 homologue of arthropods. See Hox Genes.
labial: (a) towards the periphery of the mouth (b) of or related to lips (c) a gene, abbreviated lb, the hox1 homologue of arthropods. See Hox Genes.
lamina, lamination: originally a flat metal plate (for example, the red-hot laminae ardentes used as torture device by the Romans). In biology or geology, a layer. Anything with numerous layers is thus laminated.
latilamina: in stromatoporoid anatomy, a region where the horizontal elements are closely spaced, and vertical elements may terminate, conceived as a temporary region of slow or halted growth.
lecithotrophic: (embryology) larval development in which the embryo is largely or completely non-feeding, living on stored yolk.
 lectin:
a large and important superfamily of animal proteins which bind tightly, and
very specifically, to particular sugar residues of glycoproteins.
lectin:
a large and important superfamily of animal proteins which bind tightly, and
very specifically, to particular sugar residues of glycoproteins.
lithistid: formerly, a taxonomic group of sponges, now generally regarded as polyphyletic. The lithistids have massive, silicate skeletons created by fusion of spicules. However, this appears to be a body plan which evolved several times within both the demosponges and hexactinellids. The term is sometimes used in a quasi-phylogenetic way to refer only to the demosponge lithistids.
loculus: generic anatomical term for any small compartment or recess, e.g. the spaces in the intervallum of an archaeocyath.
lophose: (sponge anatomy) of spicules, branched.
lorica: (anatomy) a hard shell, coating or crust around an organism. Maggenti et al. (2005).
MADS-box: protein structure, evo-devo. A DNA-binding protein motif present in almost all Eukarya, but particularly important in plants. See MADS box gene home page.
magnetic resonance imaging: see MRI.
mamelon: in sponge anatomy, a low, rounded protruberance, conceived of as being centered on a primary water inhalant (or, possibly, exhalant) pore.
manca: a type of isopod larva which is morphologically similar to the adult but lacks a seventh pair of pereonic appendages.
mandible: in crustacean anatomy, the most anterior pair of mouth parts, derived from the appendages of the third cephalic segment.
maxilliped: Crustacea. Anterior thoracic limb exapted to act as mouth part, with its body segment usually fused to the head (cephalon).
megasclere: (sponge anatomy), a large, typically elongate, sponge spicule.
mesohyl: (sponge anatomy) acellular layer between pinacoderm and choanoderm. The mesohyl contains spicules and amoebocytes.
metamere: in anatomy or development, a segment, usually a body segment.
metamerization: segment formation.
microbialite: ""Burne and Moore (1987) introduced the term microbialite as [meaning] organosedimentary deposits of benthic microbial communities and, following this definition, stromatolite can be seen as a type of microbialite showing lamination as a specific feature." Dupraz et al. (2006), citing Burne RV & LS Moore (1987), Microbialites: organosedimentary deposits of benthic microbial communities. Palaios 2: 241– 254 (which we have not personally read).
micromere: in embryology, the smaller of the two daughter cells from an unequal division. More generally, any small cell.
microsclere: (sponge anatomy), a small spicule.
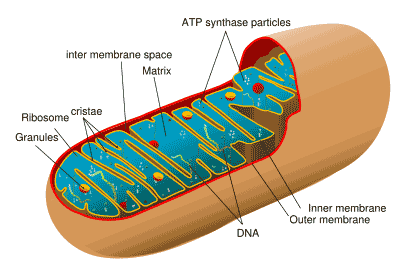 mitochondrion: an organelle responsible for most of the oxidative
metabolism in the cell. There is far too much to cover in a glossary
definition. See Mitochondrion - Wikipedia
for a relatively brief introduction.
mitochondrion: an organelle responsible for most of the oxidative
metabolism in the cell. There is far too much to cover in a glossary
definition. See Mitochondrion - Wikipedia
for a relatively brief introduction.
monactine: (sponge anatomy) a spicule or spicule axis with only one ray. See Spicule Terms.
monaxon: (sponge anatomy) a spicule with only one axis. See Spicule Terms.
mox: a class of homeobox genes, one of which is frequently located just upstream of the hox cluster (if one exists). See Hox Genes.
MRI: magnentic resonance imaging. The protons in the nucleus have a quantum mechanical property called "spin." It has nothing much to do with rotation, but it is harmless to think of it that way for our purposes. Protons normally pair up, so that net spin is zero if the nucleus has an even number of protons. However, if the element has an odd atomic number, one proton is unpaired, most significantly in hydrogen, with just one proton. exposing these atoms to an intense magnetic field forces the spins to line up (again, the analogy to an axis of rotation is false, but harmless). The target is then pulsed with electromagnetic radiation which causes some of the protons to flip. Between pulses, the protons flip back, causing a slight disturbance in the magnetic field. The wave form of this disturbance is what is actually measured. It is exquisitely sensitive to the chemical environment of the proton in hydrogen, and yeilds a detailed three-dimensional map. Unfortunately, this requires the use of Fourier transforms, which cause our eyes to cross, followed rapidly by tremors, catatonia, and eventual death. Thus we will quit now.
mu (μ): the Greek lower case mu alone, or as as "μm" signifies a micron or micrometer = 10-6 meters = 0.001 mm = 1000 nm, about the diameter of a small bacterium.
 nauplius:
(crustacean embryology)
first larval stage of crustaceans, in which the larva has three pairs of
appendages. These will later become antennae and a pair of anterior mouth
parts.
nauplius:
(crustacean embryology)
first larval stage of crustaceans, in which the larva has three pairs of
appendages. These will later become antennae and a pair of anterior mouth
parts.
neuropil: (neuroanatomy) a brain, or any large collection of neurons (ganglion), consists of cell bodies with a nucleus and all the usual machinery of metabolism, plus long cell processes which actually do the work of transmitting signals. A neuropil is a dense tangle of these processes. The cell bodies normally sit outside the neuropil, where they won't get in the way, and where they will have easier access to the glial (non-neuronal) support cells.
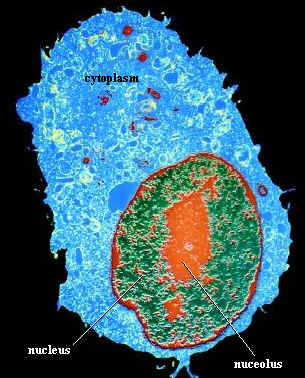 nucleolus:
(cell biology)
a dark staining body in the nucleus of many eukaryotes which is associated with
the production of ribosomal RNA.
nucleolus:
(cell biology)
a dark staining body in the nucleus of many eukaryotes which is associated with
the production of ribosomal RNA.
A B C D E F G H I J K L M N O P Q R S T U V W X Y Z
| Page Back | Page Top | Unit Home | Page Next |
Page by: ATW070701
Last Revised: ATW070825
chacked ATW070725
All text public domain. No rights reserved.