 Microsporidia (= Microspora):
Microsporidia (= Microspora):| Stem Metazoa | ||
| Eukarya | Microsporidia |
| Eukarya | Time |
| Microsporidia References |
 Microsporidia (= Microspora):
Microsporidia (= Microspora):
Range: The Microsporidia are all obligate intracellular parasites. Spores of this group appear to be nearly ubiquitous. There are currently approximately 150 described genera of Microsporidia with over 1200 individual species; and it is likely that this represents only a fraction of the total diversity of Microsporidia [KF02]. Microsporidia parasitize animals from virtually all groups (even Bryozoa! [MA02]), as well as certain other protists which are themselves animal parasites [K+01]. However, the vast majority of Microsporidia attack insects and other arthropods. Thus, the crown group of living Microsporidia is probably not much older than the Carboniferous, when insects first became common.
Phylogeny: Historically, the Microsporidia were regarded as a separate phylum of uncertain affinities [H+99]. Early molecular phylogenies using small subunit rDNA also placed the Microsporidia on a very deep branch with parabasalids and diplomonads [E+01]. As discussed below, microsporidian ribosomes are extremely aberrant [D+01], which undoubtedly accounts for this result.
The current consensus view is that Microsporidia are either the sister group of the Fungi or even a peculiar group within the Fungi. This placement is supported by molecular studies based on α and β tubulins [E+01], the transcription factor TBP [F+99], the structure and function of the microsporidian enzymes responsible for placing the 5'-cap on mRNAs [H+02], and RNA polymerase II [H+99]. EF-1α proteins of Microsporidia, animals, and Fungi all contain a unique insertion to these taxa. In addition, dihydrofolate reductase and thymidylate synthase are distinct enzymes in animals, Microsporidia, and Fungi but are united as a single enzyme in plants and other protists [K98]. Apparently, there are features of the reproductive cycle which also suggest a relationship with Fungi [K98].
 Some recent results actually place the Microsporidia well within the Fungi [K+00]. In fact, in a β-tubulin phylogeny, Microsporidia branch with either Ascomycetes or Zygomycetes, probably the two most derived of the four main fungal taxa [K+00].
Some recent results actually place the Microsporidia well within the Fungi [K+00]. In fact, in a β-tubulin phylogeny, Microsporidia branch with either Ascomycetes or Zygomycetes, probably the two most derived of the four main fungal taxa [K+00].
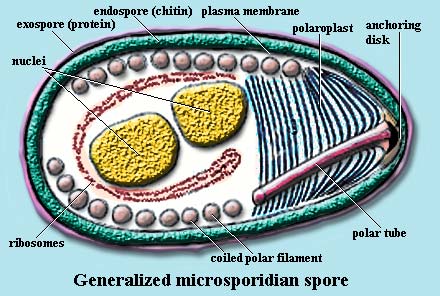
Characters: Generally small, 1-40µ long [KF02] and 0.5 - 5µ wide. The cells are usually ovate or rod-shaped.
Peripheral structures: In the spore stage, the microsporidian cell is typically protected by a 30-40 nm thick proteinaceous exospore and a 20-35 nm thick endospore layer containing chitin [B+00] and protein [H+01]. The spore coat may be somewhat simpler in other systems [C+02].
The exospore contains a protein which cross-reacts with anti-keratin antibodies, but does not seem to have significant homology with keratin, or anything else [B+00]. Apparently, like keratin, it has a C-terminal region with a repeated motif rich in glycine and serine and a number of conserved cysteine sites [H+01]. On activation this protein becomes phosphorylated and disassembles [B+00]. In Encephalitozoon intestinalis (but not two other species of Encephalitozoon), the outer coat protein is actually two proteins which are expressed at different developmental stages, with the later overlying the earlier [H+01]. The C-terminal regions of these proteins are virtually identical [H+01]. It is apparently rather common for the exospore to be made up of two distinct layers, the outer layer being frequently less tightly organized. See, e.g., [MA02] and [B+00] (characterizing the inner exospore as an "intermediate" layer). Some groups also have an outer glycocalyx in addition to the protein exospore.
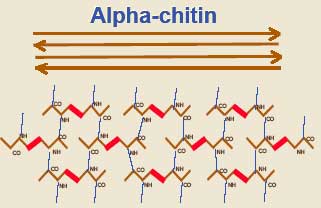
The endospore chitin is in the form of alpha chitin, the same crosslinked type thought to be a synapomorphy of Arthropoda. Given the close ecological relationship between arthropods and Microsporidia, this seems unlikely to be a coincidence. The endospore is of uniform thickness except over the anchoring disk, where it is usually significantly thinner.
Motility organs: Like all fungi except the primitive chytrids, Microsporidia lack flagella or any other "9+2" structure [FK01].
Cytoskeleton: centrosome with centrosomal plaque [K+00].
Mitochondria: Microsporidia lack mitochondria and peroxisomes [H+99]. However, they contain a heat-shock protein Hsp70) [P+98a] and several units of the pyruvate dehydrogenase (PDH) complex which appear to have been derived from a mitochondrial source [FK01]. It is uncertain how the mitochondrial PDH elements are used. However, this is in clear contrast to other "amitochondriate" eukaryotes, whose varying strategies for coping without mitochondria generally involve the substitution of PDH by pyruvate: ferrodoxin oxidoreductase [F+01] [1]. Hashimoto et al. [H+98] are of the view (and we are inclined to agree) "that no representatives of the pre-mitochondrial stage of eukaryotic phylogenesis exist among the species living today."
Stacked lamellar organelles: Stacked Golgi dictyosomes are absent [K+00].
The polaroplast takes up most of the "anterior" end of the cell. The polaroplast is a transverse structure made up of tightly folded membranes, vesicles, or both. It may also have a more loosely organized posterior region variously referred to as the "spongiform," "tubular," or "vesicular" polaroplast [D98] [C+02] [KF02].
Vacuolar organelles: At the "posterior" end of the cell, the spore usually contains a large vacuole. The function of the posterior vacuole is unknown. Like the polarosome, its principle purpose may simply be to generate the directional pressures necessary to accomplish rapid infection.
Fibrous organelles: In the spore stage, the polar filament is usually found as a series of tight coils, just below the plasma membrane. These are observed in electron micrographs as a series of dots running just under the cell membrane. The "dots" are cross-sections of the polar tube as it coils 5-15 times around the entire circumference of the spore. The number of coils, their arrangement relative to one another, and even the angle of helical tilt are conserved and diagnostic for a particular species [KF02].
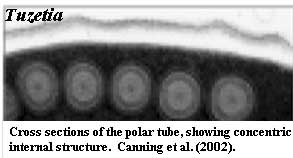 The filament ranges from 0.1 to 0.2µ in diameter and 50 to 500µ in length [KF02]. The coiled filament is surrounded by massive arrays of ribosomes, particularly in immature cells [B+02]. Anteriorly, the polar filament passes through the polaroplast and attaches to an anchoring disc at the apex of the cell. At infection, the polar filament will very quickly elongate the polar tube (up to many times the length of the spore) [FK01]. The polar tube or filament (the filament is simply an extension of the tube) is composed of membrane and glycoprotein layers [KF02] and appears to have considerable internal structure, as shown in the image of Tuzetia [C+02]. There is some physical association between the end of the polar filament and the posterior vacuole, but the precise nature and function of this contact are currently speculative [KF02].
The filament ranges from 0.1 to 0.2µ in diameter and 50 to 500µ in length [KF02]. The coiled filament is surrounded by massive arrays of ribosomes, particularly in immature cells [B+02]. Anteriorly, the polar filament passes through the polaroplast and attaches to an anchoring disc at the apex of the cell. At infection, the polar filament will very quickly elongate the polar tube (up to many times the length of the spore) [FK01]. The polar tube or filament (the filament is simply an extension of the tube) is composed of membrane and glycoprotein layers [KF02] and appears to have considerable internal structure, as shown in the image of Tuzetia [C+02]. There is some physical association between the end of the polar filament and the posterior vacuole, but the precise nature and function of this contact are currently speculative [KF02].
Two rather different proteins have been isolated from the polar tubes of Encephalitozoon. One is large (~ 50 kD) and proline-rich. The other is smaller (~30Da), with a central region containing strings of lysine and an acidic C-terminal region [D+01]. The genes encoding these proteins appear to be conserved, as is the close spacing of the two genes. Neither gene has any known homologues. Delbac et al. [D+01] have suggested that the two interact via disulfide bridges, since thiol-reducing agents (unlike most other agents) disrupt the polar tube.
Ribosomes and Protein Synthesis: Ribosomes are particularly abundant in the cytoplasm and are carried along into the infected cell with the sporoplasm. These ribosomes are 70S organelles, and not of the typical (80S) eukaryotic type [F+99] [B+02] [D+01]. In addition, the 5.8S and 28S rRNAs are fused, as they are in bacteria [F+99]. In fact, there is no spacer sequence between the two in the rDNA, and the two are transcribed as a single molecule, as in bacteria [P+98]. It was thought for some time that these ribosomes were a link with prokaryotic ribosomes. This does not appear to be the case [D+01] [P+98]. The rRNA species are far more closely aligned with eukaryotic rRNAs in both structure and sequence [P+98]. The distribution of rDNA sequences in the genome shows no pattern. In some Microsporidia, all rDNA sequences are located on a single chromosome, while in others, the rDNA sequences appear to be randomly distributed [P+98]. However, like prokaryotic ribosomes, microsporidian ribosomes contain a large component of small polyamines. Presumably, as in prokaryotes, these molecules bind to the rRNA [B+02].
Nuclei: The spore may have a single nucleus or, more typically, a diplokaryon, two nuclei in close association. The perinuclear cytoplasm is rich in ribosomes, apparently borne on endoplasmic reticulum. Microsporidia have the smallest known genomes of any eukaryote -- as little as 2.3 Mbp in one species of Encephalitozoon [D+01]. This is smaller than many bacterial genomes [FK01] (compare, e.g., E. coli with 4.6 Mbp and Homo with 3200 Mbp). Virtually all nonessential intergene regions have been deleted. In addition, for unknown reasons, the rate of mutation is extraordinarily high [K98].
 Life cycle & Reproduction: The life cycle may be simple or complex, and may involve sexual or asexual reproduction, or both. Those with complex life cycles may have multiple obligate hosts and as many as three different spore types. These forms tend to be specialized to very specific host organisms and specific tissues. Others, with simple life cycles and often asexual reproduction, have very broad tolerance and are capable, at least under laboratory conditions, of infecting nearly any type of eukaryotic cell.
Life cycle & Reproduction: The life cycle may be simple or complex, and may involve sexual or asexual reproduction, or both. Those with complex life cycles may have multiple obligate hosts and as many as three different spore types. These forms tend to be specialized to very specific host organisms and specific tissues. Others, with simple life cycles and often asexual reproduction, have very broad tolerance and are capable, at least under laboratory conditions, of infecting nearly any type of eukaryotic cell.
The usual form of the reproductive cycle is shown in the figure. The infective spores can survive for extended periods in the environment. So, for example, spores of Encephalitozoon can survive heating to 56°C for 60 min, a pH of 9 or 4 for 24 h, or storage at 4°C for 2 years without losing infectivity [H+01].When activated, the polaroplast and posterior vacuole swell rapidly due to a spike in osmotic pressure [KF02]. The anchoring disk ruptures through the thin endospore wall adjacent to it [KF02].
The spore extrudes the polar tube by eversion. That is, it turns inside-out with the dense glycoprotein core becoming an outer protective layer [KF02]. The free end of the tube inserts through the cell membrane of the host. The polar tube serves as a pliable hose through which the infectious sporoplasm is pumped into the host cell in 15 to 500 msec [KF02]. This entire process is completed in less than two seconds in the model systems in which it has been measured [D+01]. Alternatively, the spore may be internalized by phagocytosis [H+01].
The sporoplasm is probably forced through the polar tube by osmotic pressure. The spores are permeable to water and, as a result of high solute concentrations, presumably have high turgor pressures even in the inactive state. At activation, microsporidians use various means to increase this turgor, and its effectiveness. In one, well-studied system, the glucose disaccharide, trehalose, is rapidly hydrolyzed to glucose. Since osmotic pressure depends on the number of solute molecules, and not their mass, this results in a sharp spike in pressure. Likewise, other species may rapidly flood the cytoplasm with calcium ions, which has precisely the same effect.
The sporoplasm consists of the nuclei and surrounding cytoplasm. Microsporidian ribosomes are a large component of the sporoplasm; and these ribosomes promote a very high rate of protein synthesis during the initial infective cycle [B+02]. When the sporoplasm emerges from the tube, it has somehow already acquired a new cell membrane. This is thought to derive from elements of the polaroplast which precede the sporoplasm through the tube [KF02]. Inside the host cell, the nuclear material in the sporoplasm replicates extensively, either in direct contact with the host cytoplasm or inside a parasitophorous vacuole.
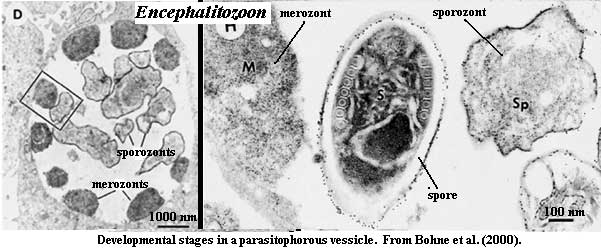 Although there is considerable variation, a typical microsporidian may replicate by merogony for some initial period, once it is inside the host cell. During this period, in some cases, the nuclei may proliferate with or without division into individual cells [C+02]. Within the first 24-48 hours after the sporoplasm has reached the host cell, several rounds of division have occurred.
Although there is considerable variation, a typical microsporidian may replicate by merogony for some initial period, once it is inside the host cell. During this period, in some cases, the nuclei may proliferate with or without division into individual cells [C+02]. Within the first 24-48 hours after the sporoplasm has reached the host cell, several rounds of division have occurred.
Synthesis of the spore coats and sporogony then begin [B+02]. In species which reproduce within a vacuole, the initial steps in replication take place in close association with the inner membrane of the vacuole. The transition to sporogony is marked by release of the developing spore into the lumen of the vacuole [B+00] and the accumulation of electron dense material near the periphery of the cell [H+01]. Both merozonts and sporozonts show little internal organization [H+01] (see also images from [B+00]). In at least two, widely divergent systems, electron-dense extracellular tubules have been observed surrounding developing spores during sporogony [B+00] [C+02]. When the spores completely fill the host cell cytoplasm, the cell lyses and releases the spores to the surroundings. In some systems, the microsporidian infestation may cause the development of xenomas. See, e.g., [MA02].
Habitat & ecology: The same organism may have several different spore types. For example, different spores may be produced on infection of primary and secondary hosts, and spores designed primarily for internal infection of additional host cells may differ from those specialized for survival in the environment.
One microsporidian, Nosema locustae, is even commercially marketed (as NoLo Bait) for biological control of grasshoppers, locusts and crickets. However, a related species, Nosema apis, is a serious problem for bee keepers.
Images: Enterocytozoon life cycle from the Atlas of Medical Parasitology of the Carlo Denegri Foundation;
Links: Protozoa and Microsporidia discusses possible use in agriculture for pest control); SCSB #387 - Microsporidia (Protozoa)- A Handbook of Biology and ... wonderful on-line resource with much hard information); DPDx - Microsporidiosis a good, quick explanation of the life cycle). Biology of Microsporidia.
References: Bacchi et al. (2002) [B+02], Bohne et al. (2000) [B+00], Canning et al. (2002) [C+02], Delbac et al. 2001) [D+01], Didier (1998) [D98], Edgcomb et al. (2001) [E+01], Fast & Keeling (2001) [FK01], Fast et al. (1999) [F+99], Hashimoto et al. (1998) [H+98], Hausmann et al. 2002) [H+02], Hayman et al. (2001) [H+01], Hirt et al. (1999) [H+99], Keeling (1998) [K98], Keeling & Fast (2002) [KF02], Keeling et al. (2000) [K+00], Morris & Adams (2002) [MA02], Peyretaillade et al. (1998) [P+98], Peyretaillade et al. (1998a) [P+98a].
Notes: [1] This is the enzyme also characteristic of hydrogenosomes [K98].