|
|
References |
| Bacteria |
References |
A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z
Cavalier-Smith, T (2002), The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int'l. J. Syst. Evol. Microbiol. 52: 7–76.
Creevey, CJ, DA Fitzpatrick, GK Philip, RJ Kinsella, MJ O'Connell, MM Pentony, SA Travers, M Wilkinson & JO McInerney (2004), Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc. Roy. Soc. Lond. B 271: 2551–2558.
Farr, GW, K Furtak, MB Rowland, NA Ranson, HR Saibil, T Kirchhausen & AL Horwich (2000), Multivalent Binding of Nonnative Substrate Proteins by the Chaperonin GroEL. Cell 100: 561–573.
Finbow, ME & MA Harrison (1997), The vacuolar H+-ATPase: a universal proton pump of eukaryotes. Biochem. J. 324: 697–712.
Garrity, GM & JG Holt (2001), The road map to the Manual, in DR Boone & RW Castenholz [eds.] Bergey's Manual of Systematic Bacteriology, 2nd ed. Springer 1: 119–166.
Horn, C, B Paulmann, G Kerlen, N Junker & H Huber 1999), In vivo observation of cell division of anaerobic hyperthermophiles by using a high-intensity dark-field microscope. J. Bacteriol. 181: 5114–5118.
Kimura, M & T Ohta (1974), On some principles governing molecular evolution. Proc. Nat. Acad. Sci. (USA)
71: 2848–2852.
Lodes, MJ, Y-Z Cong, CO Elson, R Mohamath, CJ Landers, SR Targan, M Fort & RM Hershberg (2004), Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Invest. 113: 1296–1306.
Miroshnichenko, ML, NA Kostrikina, NA Chernyh, NV Pimenov, TP Tourova, AN Antipov, S Spring, E Stackebrandt & EA Bonch-Osmolovskaya (2003), Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Intern. J. Sys. Evol. Microbiol. 53: 323–329.
Nagy, I, T Tamura, J Vanderleyden, W Baumeister, & R DeMot (1998), The 20S proteasome of Streptomyces coelicolor.
J. Bacteriol., 180: 5448–5453.
Oster, G, H-Y Wang & M Grabe (2000), How F0-ATPase generates rotary torque. Phil. Trans. Roy. Soc. Lond. B 355: 523–528.
Pace, NR (1997), A molecular view of microbial diversity and the biosphere. Science 276: 734–740.
Penny, D & A Poole (1999), The nature of the last universal common ancestor. Curr. Op. Genet. Devel. 9: 672–677.
Philippe, H & P Forterre (1999), The rooting of the universal tree of life is not reliable. J. Mol. Evol. 49: 509–523.
Ranson, NA, HE White & HR Saibil (1998), Review: Chaperonins. Biochem. J. 333: 233–242.
Suhre, K, J Navaza & Y-H Sanejouand (2006), NORMA: a tool for flexible fitting of high-resolution protein structures into low-resolution electron-microscopy-derived density maps. Acta Cryst. D62: 1098–1100.
Takagi, F, N Koga & S Takada (2003), How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: Molecular simulations. Proc. Nat. Acad. Sci. (USA) 100: 11367–11372.
Thirumalai, D, DK Klimov & GH Lorimer (2003), Caging helps proteins fold. Proc. Nat. Acad. Sci. (USA)
100: 11195–11197.
Woese, CR (2002), On the evolution of cells. Proc. Nat. Acad. Sci. (USA) 99: 8742–8747.
Woese, CR, O Kandler & ML Wheelis (1990), Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Nat. Acad. Sci. (USA) 87: 4576–4579.
[1] Where the term 'morphological' characters appears in single quotes, it is used in a broader sense than usual—not just morphological characters per se, but any character that might arguably be used in a phylogenetic analysis the same way as a true morphological character. So synthesis or not of certain proteins, or gene-structural characters such as indels, might also count as 'morphological' characters.
[2] Here's the remedial thermodynamics for those who need it. The magic equation is ΔG = ΔH - TΔS. The free energy (ΔG) needed to get something done at constant temperature is equal to the change in potential energy (enthalpy) between the initial and final states (ΔH) less the absolute temperature multiplied by the change in entropy (TΔS). Entropy is an elegant philosophical and and physical concept which can variously be conceptualized as "disorder" or "randomness," but also "freedom" or "potential number of future states." We pause briefly to allow the philosophical implications to sink in... Ready? Now, consider a simple model of a protein as a chain of paperclips. We know that the final, properly folded state of the protein has low enthalpy because the final state is quite stable. It has to be or the protein could never maintain its useful shape. So ΔH is negative, and the reaction is favored. So far so good. However, if we have to fasten together distant parts of the chain, this drastically reduces the number of states which the chain can assume. This reduces entropy, and the -TΔS term becomes strongly positive, especially under heat stress (higher T), and the conformation change becomes prohibitively expensive in terms of free energy. When the chain is suckered into a small volume, taking due advantage of random fluctuations in shape to accomplish the task, the number of possible conformations is strongly reduced because there is no room to assume extended shapes. Thus the change in entropy needed to make distant parts of the chain interact is no longer so great a problem, and the final state is reached without much grief. (This sounds as if we're getting something for nothing, and reducing total entropy, neither of which happen in thermodynamics. The key is that the GroEL molecule and ATP have to be considered as part of the system.)
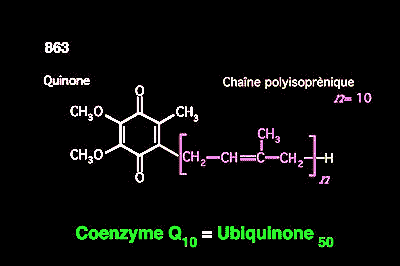 [3] c'est-à-dire un alcool avec une longue chaîne composées de plusieurs (une dizaine) unités de 3-mèthyl-2-butene (isoprène). Ça c'est un motif chimique très courrant, par exemple en l'omniprésent co-facteur métabolique ubiquinone. Image: Faculté de la Pitié Salpêtrière.
[3] c'est-à-dire un alcool avec une longue chaîne composées de plusieurs (une dizaine) unités de 3-mèthyl-2-butene (isoprène). Ça c'est un motif chimique très courrant, par exemple en l'omniprésent co-facteur métabolique ubiquinone. Image: Faculté de la Pitié Salpêtrière.
checked ATW??????, edited RFVS111214
 [3] c'est-à-dire un alcool avec une longue chaîne composées de plusieurs (une dizaine) unités de 3-mèthyl-2-butene (isoprène). Ça c'est un motif chimique très courrant, par exemple en l'omniprésent co-facteur métabolique ubiquinone. Image: Faculté de la Pitié Salpêtrière.
[3] c'est-à-dire un alcool avec une longue chaîne composées de plusieurs (une dizaine) unités de 3-mèthyl-2-butene (isoprène). Ça c'est un motif chimique très courrant, par exemple en l'omniprésent co-facteur métabolique ubiquinone. Image: Faculté de la Pitié Salpêtrière. 