
All material by ATW may be used under the terms of a
Public Domain Dedication.
| Glossary | ||
| Bacteria | Glossary A–Z |
| Bacteria | Bacteria | Time |
| LUCA
├─Eubacteria
│ ├─Actinobacteria
│ └─┬─Firmicutes
│ └─Didermata
│ ├─Cyanobacteria
│ └─┬─Sphingobacteria
│ └─Proteobacteria
└─Neomura
├─Archaea
└─Eukarya
├─Chlorobionta
└─┬─Fungi
└─Metazoa
├─Deuterostomia
└─Protostomia
|
Lists |
Active transport transport across a cell membrane which requires energy generated by the hydrolysis of some energy carrier (usually ATP).The selective active transport of ions (usually sodium) out of the cell is often used as a secondary energy storage mechanism. See ion gradient system.
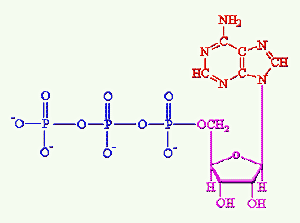
ATPadenosine triphosphate. This is the common currency of chemical energy in most cells. It is more or less accurate to say that the whole object of metabolizing food, of whatever kind, is to recover energy to make ATP. Specifically, the energy from the breakdown of food is used to add a third phosphate group onto adenosine diphosphate (ADP) to make the triphosphate. The energy stored in ATP is then used to drive various reactions by cleaving the third phosphate back off (hydrolysis of ATP). This job is done by enzymes (ATPases) which couple the ATP hydrolysis with some other reaction which absorbs energy. Chemically, ATP is built from exactly the same chemical unit that supplies the adenosine or "A" monomers to RNA. Now, stop fidgeting and pause a moment to reflect on the evolutionary implications of all this: precisely the same molecule, phosphorylated adenosine, is at the core of (a) most information exchange (b) the vast majority of food metabolism reactions, and (c) most synthetic reactions, in every known organism. Why? No one knows the answer to this question. This is not the sort of molecule you'd expect to form spontaneously—or at least not under the kinds of conditions found today. There are two relatively high-probability implications. (1) We may be missing a lot of hidden evolution—lost diversity—which occurred between the first living organism and the last common ancestor of all organisms alive today. (2) Life may have gotten started at the interface between radically different chemical environments. Notice that the adenosine base in ATP is strongly reduced, non-polar, compact, and basic. The phosphate end is highly oxidized, ionic, has an extended shape, and is quite acidic. The ribose ring in between is intermediate in all four respects. It's hard to see how this molecule could form without either a sophisticated biochemical system already in place (implication "a"), the close juxtaposition of wildly different chemical environments (implication "b"), or both.
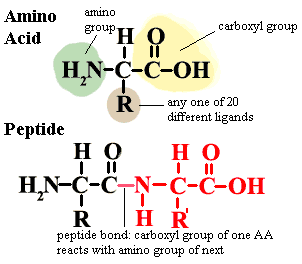
Amino acid the fundamental building block of proteins. There are twenty different amino acids normally found in proteins. All have the general structure shown in the figure. In proteins, the amino acids are joined by peptide bonds as shown in the image. Notice that the central carbon atom has four different ligands. It is therefore asymmetrical and can exist in two mirror image forms (enantiomers), known as L and D enantiomers. Proteins in living organisms are all made from L-amino acids. However bacterial cell walls and a few other structures incorporate some D-amino acids. A few naturally occurring amino acids are not normally found in proteins and are not specified in the genetic code. Ornithine (R = (CH2)3NH2) is one example. These are non-protein amino acids are common intermediates in a variety of metabolic pathways. Finally, some amino acids may be chemically modified after they have been incorporated into proteins.
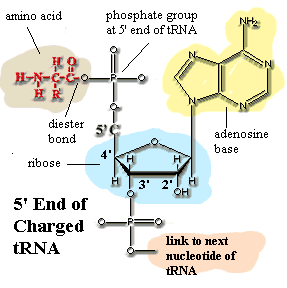
amino acyl-tRNA a "charged" tRNA, i.e. one with a bound amino acid. The carboxyl group of the amino acid is bound through a diester linkage to the 5' phosphate ligand of a terminal adenine on the tRNA. This leaves the amino group free to form a peptide bond with the growing protein chain on a ribosome. The amino-acyl tRNA for alanine may be abbreviated ala-tRNA, tRNAala, or even ala-tRNAala. Generally, the subscript refers to the tRNA species, and the superscript shows what it is actually charged with.
amino group a ligand of the form -NH2.
anticodon the complementary RNA or DNA sequence to that of a codon. Thus, GCG is one of the codons for alanine. See genetic code. The corresponding anticodon would be CGC.
Apomorphy a character state which is unique to a single, terminal taxon. Example: among primates, complex grammar is an apomorphy of human beings. It is quite diagnostic of humans, but useless in determining phylogenetic relationships because it is not a shared, derived characteristic, or synapomorphy, of any larger group.
Autotroph an organism which obtains energy from inorganic sources, sunlight or the oxidation of inorganic chemicals.
Autotrophic nutrition synthesis of organic food molecules from inorganic compounds such as carbon dioxide.
carboxyl group a ligand of the form -COOH, i.e., a simple organic acid.
Chaperonin any of a class of ATP-dependent (i.e. they need chemical energy to do the job) proteins which are responsible for folding polypeptides into the correct conformation. As anyone who has been confronted with a spool of ribbon and a pile of presents knows, the ribbon will not fold itself into an appropriately decorative conformation. Even though the desired conformation is stable and energetically favorable, the ribbon needs energetic guidance to attain this state within some biologically relevant timescale. This is the function of chaperonins. They are composed of two doughnut-shaped subunits. Chaperonins also have a limited ability to "repair" proteins which have been incorrectly folded. Class I chaperonins are chaperonins closely related to the E. coli GroEL protein, and are sometimes referred to as GroE chaperonins. The holoenzyme is composed of two heptameric subunits and works in concert with a helper Hsp 10 protein (GroES). Class I chaperonins are found in Eubacteria, mitochondria, and chloroplasts. Class II chaperonins, or TCP1 proteins include both the thermosomes or TF55 proteins of Archaea and the CCT proteins of eukaryotes. The eukaryotic species are composed of two octameric rings, while thermosomes may have either 8 or 9-member rings. Class II chaperonins do not have a helper protein. Class II chaperonins are very closely related to Class I chaperonins by structure, but not by sequence. For more information, see Pieces: GroEL.
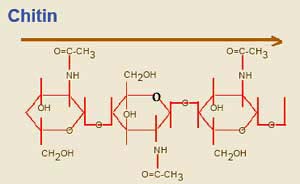
Chitin a polymer of repeating sugar molecules (a slightly modified glucose, poly-N-acetyl-D-glucosamine). See image. Chitin is the material which makes up the exoskeleton of insects and, in more or less modified form, in almost all arthropods. In arthropods, chitin occurs in a crosslinked form, α-chitin. Significantly, it is also found in the radular "teeth" of molluscs, the setae (bristles) and jaws of annelid worms, and the cell walls of Fungi. So, this is exceedingly ancient stuff, possibly predating the split between bacteria and metazoans.
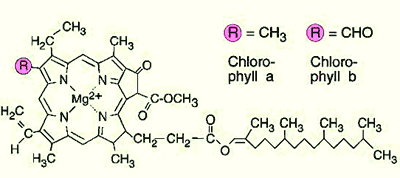
chlorophyll a widely dispersed photosynthetic pigment, particularly effective in red and blue light (it reflects the mid-range green wavelengths, which is why it appears green to our eyes). Note that chlorophylls a and b differ only in the substitution of a methoxy for a methyl ligand in one position.
Clade a group of organisms consisting of an organism and all of its descendants.
Codon the basic element of the genetic code. A sequence of three nucleotides that specifies a particular amino acid, or serves as a "start" or "stop" signal for translation.
Crista pl. cristae) (1) of mitochondria, folds in the internal membrane of the mitochondrion which gives the organelle its characteristic appearance. This is the site of the electron transport chain in oxidative metabolism. The cristae, therefore, serve as the physical link between the tricarboxylic acid cycle and oxidative phosphorylation (ATP synthesis). See also Wikipedia: Mitochondrion. (2) more generally, crest (its literal meaning in Latin) or ridge.
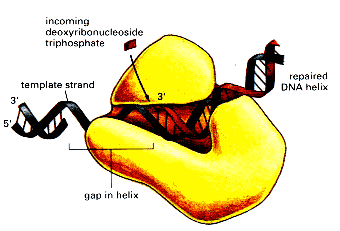
DNA polymerase an enzyme which replicates DNA, either as a part of cell replication or DNA repair.
DNA polymerase III The dominant DNA polymerase in E. coli. By extension, similar DNA polymerases in other Eubacteria (i.e., Type C polymerases) are often referred to by the same name. "DNA Polymerase III is an asymmetrical dimer, composed of 18 subunits, a complex arranged from combination of 10 distinct subunits. Three subunits form the core of the enzyme, these are alpha, epsilon and theta. The holoenzyme is created from other subunits (beta, delta, delta prime, chi, gamma, psi, and tau) variationally binding to this core, and conferring the full functions and characteristics the enzyme needs to carry out the replication of DNA. As a replicative enzymatic mechanism of DNA, the Polymerase replicates with high fidelity. To maintain this level of fidelity, a proofreading mechanism has been included, by evolution, in the enzyme, in the form of its epsilon subunit." DNA Polymerase III. Image from the same page.
DNA polymerase, Type C: DNA polymerases with sequence and structure similar to E. coli polymerase III. Type C polymerases are restricted to Eubacteria. By sequence they have no detectable relationship to polymerases from Archaea or eukaryotes.
DNA topoisomerase any of a class of enzymes that alter or maintain the topological structure of DNA, e.g., in a supercoiled form.
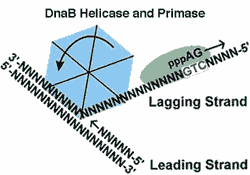
DnaB a bacterial helicase. There are several helicases, but this is the main one involved in DNA replication in Eubacteria. It works cooperatively with DNA polymerase III to unwind DNA ahead of DNA synthesis on the leading strand (i.e., 5' → 3'). Every 500 or 100 nucleotides, it stimulates primase to create an RNA primer for replication of the lagging strand. DnaB is an ATP-dependent homohexamer. See DNAB HELICASE HOME PAGE, also the source of the figure.
DnaK a bacterial chaperonin homologous to the Hsp70 of eukaryotes. DnaK works in tandem with DnaJ and GrpE to accomplish ATP-dependent folding of polypeptides. DnaK is a heat shock protein. However, when not operating under heat shock conditions, DnaK is also present and is involved in the degradation of σ32, an RNA polymerase regulator which detects the promoter sites for transcription of RNA coding for heat shock proteins.
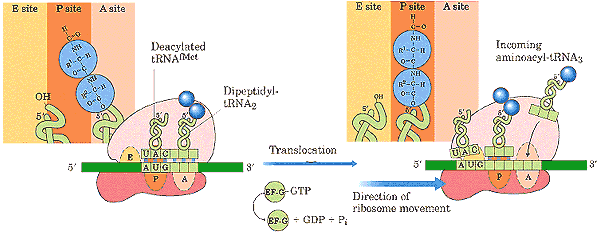
EF elongation factor, q.v.
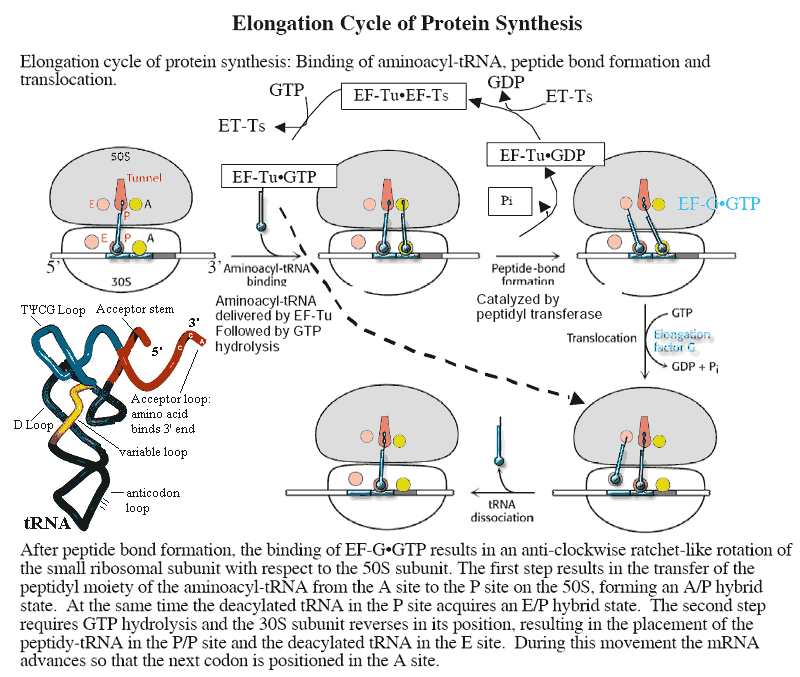
EF-G the elongation factor responsible for moving peptidyl-tRNA from the ribosomal A-site to the P-site during translation. Like EF-Tu, it makes use of a ribosomal GTPase to rotate the small (30S) ribosomal subunit with respect to the large (50S) subunit. EF-G is homologous to the EF-2 of eukaryotes.
EF-Tu the elongation factor responsible for attachment of an incoming amino acyl-tRNA to the ribosomal A-site. Like EF-G, it makes use of a ribosomal GTPase to rotate the small (30S) ribosomal subunit with respect to the large (50S) subunit. EF-Tu is homologous to the EF-1α of eukaryotes.
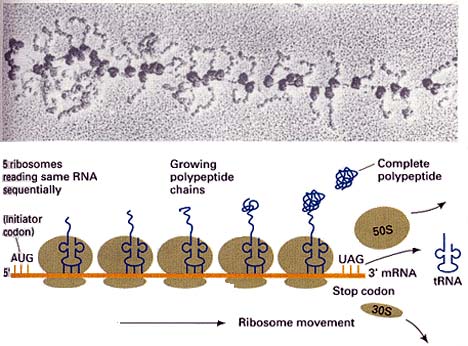
Elongation Factor during translation of mRNA on ribosomes, each tRNA binds successively to three sites on the ribosome: the A (acceptor) site, the P (peptide site), and the E (exit) site. Elongation factors are small, GTP-dependent proteins which are instrumental in this process. EF-Tu (homologous to EF-1α of eukaryotes) binds the tRNA into the A site. After the amino acid on the tRNA has been added to the growing peptide chain, EF-G (homologous to EF-2 of eukaryotes) is responsible for moving the peptidyl-tRNA complex to the P site. EF-Tu and EF-G are themselves believed to be homologous. That is, they share sequences which suggest that the genes coding for these proteins derived from a single common ancestral gene, probably before LUCA. The operation of elongation factors is described in a bit more detail in the figure at tRNA.
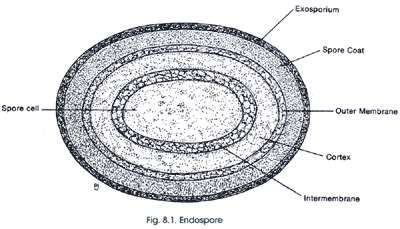
endospore the nearly immortal resting state of some bacteria. Endospores are composed of a central spore cell, which is surrounded by various protective layers. The outermost layer is the exosporium, which is a thin covering made of protein. Below this is the spore coat which is made up of highly cross-linked keratin and layers of spore-specific proteins. The cortex consists of loosely cross-linked peptidoglycan. The innermost spore cell contains the components of the vegetative bacterial cell (the cell wall, cytoplasmic membrane, cytoplasm, nucleoid, etc.). The water content of endospores is only about 10–30% of the water content of vegetative cells; therefore, endospores are capable of surviving at levels of dehydration that would kill vegetative cells. The low water content also provides the endospore with chemical resistance (to chemicals such as hydrogen peroxide) and it causes the remaining enzymes of the spore cell to become inactive. One chemical produced by endospores that is thought to lend to their high resistance is dipicolinic acid. This chemical has been found in the spore cell of all endospores examined. Dipicolinic acid interacts with calcium ions to form calcium dipicolinate, which is the main substance believed to lend endospores their resistance and represents about 10% of the dry weight of an endospore. The spore cell also contains small acid-soluble spore proteins (SASPs). These function to protect DNA from UV radiation, dessication and dry heat, and they also serve as a carbon and energy source during the germination process (conversion back to a vegetative cell). Another component of endospores that contributes to their resistant to chemical agents is the strong spore coat, which is composed of highly cross-linked keratin. Identification of particular organisms can be aided by the presence, location and size of endospores. Endospores can be located centrally, terminally or subterminally within a cell. Sometimes the endospore is much larger in diameter than the cell, which causes the cell to appear swollen at the location of the endospore." Endospore Structure.
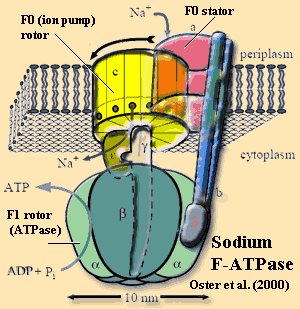
F-ATPase a very ancient, but sophisticated, molecular machine found in Eubacteria, mitochondria, and, in modified form (V-ATPase, A-ATPase, etc.), in all organisms. Very generally, an F-ATPase consists of a rotating ion pump (F0) coupled by a shaft to an ATPase (F1). In Eubacteria, the F-ATPase normally runs in "reverse," allowing ions (usually H+, sometimes Na+) at high concentration to enter the cytoplasm, driving ATP synthesis. The mechanism has been described as a "Brownian ratchet." It works a little like an old-fashioned coin-operated vending machine. The ion enters a dead end "coin slot" between the stator element and the rotor element of the ion pump. Random thermal motions of the rotor (since there is no hand to turn a crank) align the slot with a series of polar amino acid groups on the lower half of the rotor. The ion drops through into this region, which blocks the rotor from rotating backwards (hence the "ratchet") as well as blocking this lower half of the slot. When the next ion enters the upper slot and the rotor turns again, the first ion is released into the cytoplasm. This creates a net torque on the shaft which rotates the F1 ATPase, driving ATP synthesis. Free energy calculations suggest that this process is nearly 100% efficient. See mini-review by Oster et al. (2000).
flagellin a fibrous protein which forms most of the shaft in the flagellae of prokaryotes. This is quite different from the 9+2 microtubular rosette which makes up the core of eukaryotic flagellae. Interesting fact: an aberrant inflammatory response to flagellin turns out to be one of, and perhaps the main causative agent in Crohn's Disease. Lodes et al. (2004).
fmet abbreviation for N-formyl methionine, q.v.
formyl a general term for a ligand consisting of a one-carbon organic acid (carboxyl group), based on formic acid, HCOOH. The nomenclature is ambiguous, since the same term is often used to mean any of the following: HCOO-R, HCO-R, and even (wrongly) R-CO-R' (instead of keto) or R-COOH (instead of carboxy).
|
T (or U) |
C |
A |
G |
|---|---|---|---|---|
T (or U) |
TTT Phe (F) |
TCT Ser (S) |
TAT Tyr (Y) |
TGT Cys (C) |
C |
CTT Leu (L) |
CCT Pro (P) |
CAT His (H) |
CGT Arg (R) |
A |
ATT Ile (I) |
ACT Thr (T) |
AAT Asn (N) |
AGT Ser (S) |
G |
GTT Val (V) |
GCT Ala (A) |
GAT Asp (D) |
GGT Gly (G) |
genetic code the standard code is shown in the table. Each sequence of three nucleotides in DNA or RNA potentially specifies an amino acid. In RNA, all T (thymidine) bases are replaced by U uracil). Other than this, the DNA and RNA codes are the same. During translation, ribosomes and associated enzymes "read" mRNA containing the code and assemble chains of amino acids (i.e. proteins) according to this blueprint. The code is redundant, in that each amino acid (except tryptophan and methionine) is specified by more than one series of codons nucleotide bases). The sequences UAA, UAG, and UGA signal the ribosome to terminate translation. There are minor variations in the code. However, exceptions to the standard code are very rare.
glycocalyx a typically loose extracellular layer of polysaccharides.
GroEL a chaperonin of Eubacteria. For more information, see Pieces: GroEL.
GroES a helper protein of the chaperonin GroEL. For more information, see Pieces: GroEL.
heat shock proteins a distinct population of proteins synthesized under conditions of heat stress. RNA polymerase does not usually recognize the promoter sites for these proteins on the bacterial DNA. This requires a special "sigma factor," σ32. Sigma 32 is produced constitutively by the cell but, normally, is selectively degraded with a half life of about 60 sec. Under heat stress conditions, degradation is suppressed, and σ32 binds to RNA polymerase, allowing it to recognize these promoters and to synthesize mRNA coding for the heat shock proteins.
helicase an enzyme which unwinds and separates the two strands of DNA for repair or replication.
Histones: a family of strongly conserved, highly basic (arginine and lysine-rich) DNA-binding proteins found in eukaryotes and Euryarcheota. Among other functions, histones maintain DNA in a supercoiled form and act as a sort of universal repressor for regions of the genome which do not have to be immediately available for transcription.
Holoenzyme a fully-assembled enzyme in functional form, including all polypeptides and subunits. Many enzymes function as n-mers (typically dimer or tetramer). In those cases, the holoenzyme includes the entire n-mer.
Hsp see heat shock proteins.
Hsp10 the polypeptide monomer of GroES.
Hsp60 (a) the polypeptide monomer of Class I chaperonins. b) any Class I chaperonin.
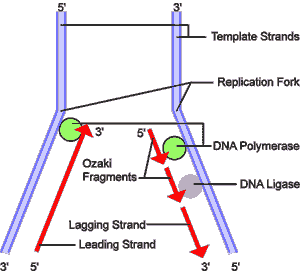
lagging strand DNA replication by DNA polymerase requires a short RNA primer sequence and works only in one direction, 5' to 3'. Since the two strands of DNA are antiparallel, this creates a problem. As a DNA helicase separates and unwinds the DNA strands, on one strand, the leading strand, the separated strand DNA strand is exposed 3' to 5'. Synthesis of a new antiparallel strand can then proceed from a new 5' end, and follow the helicase without interruption. However, on the other strand, the lagging strand, DNA synthesis must proceed in the reverse direction. This is accomplished by a secondary feature of the helicase. Every 500 or 1000 nucleotides, the helicase stimulates a primase to nick the lagging strand and insert a few RNA nucleotides as a primer. The DNA polymerase then proceeds to replicate that strand—still 5' to 3' but now in the opposite direction because it is working on the opposite strand of the original DNA. Replication of the lagging strand continues until the polymerase reaches the last point at which the lagging strand was replicated. A DNA ligase then removes the primer from this previous fragment (an Ozaki fragment), and zips the two Ozaki fragments together.
ligand a functional group in a molecule. The term usually refers to something relatively small and straightforward, e.g., a carboxyl group, a phenyl group.
LUCA Last Universal Common Ancestor. The last common ancestor of all extant species: Eubacteria, Archaea, and Eukarya.
mRNA: RNA species which are used as templates to produce proteins (in the process known as translation). Prokaryotic mRNA consists of an unbroken series of nucleotide bases in the triplet genetic code for amino acids.
murein the cell wall material of Eubacteria, a/k/a peptidoglycan. The structure of murein is discussed here.
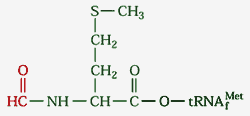
N-formyl methionine usually abbreviated fmet, an amino acid derivative used, together with a special tRNAf, to initiate translation. tRNAf bears the same anticodon as the tRNA for methionine. However, tRNAf has unique structural features, such as the sequence GCGCGC on the anticodon stem. See Protein Synthesis: The Ingredients, also the source of the image. fmet is formed from methionine by a specific formyl transferase, using formyl tetrahydrofolate as the formyl group donor. fmet is used only in protein initiation. When the polypeptide translation is complete, fmet is cleaved off. Our understanding is that this occurs in two steps, with the formyl group removed first, then the methionine.
peptide bond an amide linkage between two amino acids. See, amino acid.
peptidoglycan the cell wall material of Eubacteria, a/k/a murein. The structure of murein is discussed here.
photosynthesis a process by which an organism uses energy from light and an (usually) inorganic electron source to reduce organic compounds. There are three major groups of photosynthetic bacteria: cyanobacteria, purple bacteria, and green bacteria. The cyanobacteria carry out oxygenic photosynthesis, that is, they use water as an electron donor and generate oxygen during photosynthesis.The photosynthetic system is located in an extensive thylakoid membrane system that is lined with particles called phycobilisomes. The green and purple bacteria carry out anoxygenic photosynthesis. They use reduced molecules such as H2, H2S, S, and organic molecules as an electron source and generate NADH and NADPH. In green bacteria, the photosynthetic system is located in ellipsoidal vesicles called chlorosomes that are independent of the cytoplasmic membrane. In purple bacteria, the photosynthetic system is located in spherical or lamellar membrane systems that are continuous with the cytoplasmic membrane. Introduction to Photosynthesis.
polypeptide any relatively long chain of amino acids linked by peptide bonds. Usually treated as synonymous with "protein," however the term polypeptide is not restricted to structures similar to those produced by organisms.
protease an enzyme evolved to digest other proteins into their component amino acids.
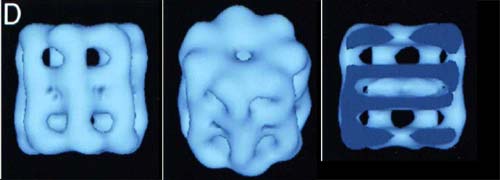
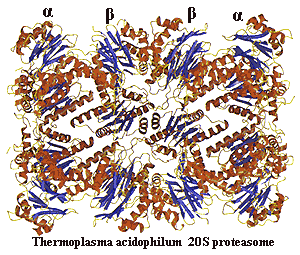
proteasome a proteasome is the evil twin of a chaperonin. Like a chaperonin, it is a longitudinally symmetrical series of 7-member rings but four rings, not two) with a hole through the middle and an affinity for improperly-folded polypeptides. Unlike a chaperonin, a proteasome does not restore proteins to conformational health and send them on their way. Instead, the proteasome slashes the doomed polypeptide into little 7–13 amino acid oligopeptides so that they may be completely degraded by other proteases. Since its function is reclamation, not rehabilitation, a proteasome lacks any of the bells and whistles of a chaperonin and possesses a much narrower aperture. Thus, only proteins which are more or less completely unfolded can pass into the destructive core of the complex. The basic 20S proteasome is found in Actinobacteria and Archaea. In eukaryotes, the 20S structure forms the core of a 26S proteasome. Image from the Max-Planck-Institut für Biochemie, Abteilung für Strukturforschung.
ribosomal RNA ribosomes are the small, but incredibly complex nucleoprotein complexes responsible for protein synthesis. They bind to mRNA molecules from the nucleus and physically move along the molecule, "reading" the code on the mRNA and attaching amino acids to the growing peptide (protein) chain. In eukaryotes there are three, quite distinctive RNA species bound up in the ribosome. These are known by their "Svedberg" numbers, a possibly obsolete measure of relative movement in centrifugation through a density gradient. The three species in prokaryotes are the 5S, 16S and 23S RNAs, vide infra.
ribosomal RNA, 16S or ssu RNA: the RNA molecule associated with the small ribosomal subunit. The secondary structure of typical 16S rRNA is extremely complex. An exhaustive set of maps and sequences can be found at rRNA secondary structure models.
ribosome the cellular organelle responsible for translating mRNA into protein. Ribosomes are complexes of specialized RNA species and numerous proteins. A really outstanding short explanation of the ribosome can be found at Ribosome Structure and Function.
RNA polymerase any of the enzyme complexes directly responsible for transcription—the manufacture of RNA from the DNA template.
rRNA ribosomal RNA, q.v.

Sliding clamp: the usual toroidal form of the DNA replication complex, including (in Eubacteria) two copies of DNA polymerase III, DNA topoisomerases, and associated proteins. Image from the von Hippel Lab at the University of Oregon.
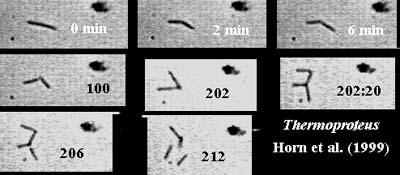
snapping division: a mode of cell division characterized by rapid horizontal division of an elongated cell with the two daughters finishing at an angle to each other. The daughter cells frequently remain in contact for a period of time. The image shows two rounds of snapping division in Thermoproteus, an archaean, in time lapse photography. From Horn et al. (1999).
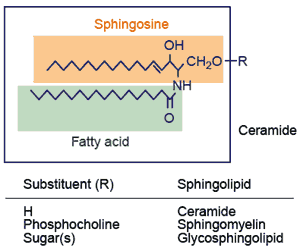
sphingolipid "All sphingolipids contain a sphingoid long-chain base (e.g. sphingosine) that is linked to a fatty acid molecule through an amide bond, thereby forming the ceramide unit. Addition of phosphocholine or carbohydrates to ceramide leads to sphingomyelin or glycosphingolipids, respectively." General sphingolipid structure.
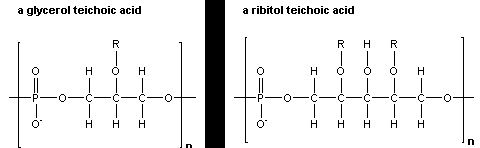
teichoic acid anionic, phosphate-rich polymers linked to the peptidoglycan of gram-positive bacteria. "They are made up primarily of repeating units of either glycerophosphate or ribitol phosphate molecules that have sugars and D-alanine attached to the glycerol or ribitol backbone. Some teichoic acids are attached to membrane lipids and these are called lipoteichoic acids. All gram-positive organisms contain lipoteichoic acids but some may lack the peptidoglycan bound form." Dr. Stuart Hill's Bios 213 Lecture notes, Lecture 9.
thylakoid a unit of a stacked, lamellar membrane system in most cyanobacteria on which photosynthesis is carried out.
transfer RNA see tRNA.
translation the process whereby the genetic code carried by mRNA is read and used to construct proteins. This process is carried out by ribosomes. The ribosomes recruit appropriate 4S or transfer RNAs (tRNAs) which are conceptually) molecules with an amino acid at one end and an "anticodon" at the other. The anticodon consists of three nucleotide bases which are the complement of the codon which codes for the tRNA's amino acid. Thus, for example, proline is coded by the sequence CCA. The corresponding tRNApro would then bear a proline amino acid at one end, and the complementary sequence, i.e. GGU, at the other. The ribosome sits on the mRNA molecule. If the ribosome detects that the tRNA bases form complementary base pairs with the next mRNA triplet in line, it clips the amino acid off the tRNA and ads it to the growing protein. It then moves up three bases on the mRNA and looks for the next matching tRNA. See also image at tRNA.
tRNA 4S RNA. This is the RNA species which actually makes the connection between genetic code and amino acid. There are 61 different tRNA species—one for each of the 64 possible codons except the termination signals. Each tRNA has an anticodon segment, with three exposed bases complementary to a particular codon. Each tRNA can be "charged" with a particular amino acid by a specific amino acyl-tRNA transferase. During translation, the tRNA which bears the appropriate anticodon base pairs with the mRNA which is being translated. The tRNA is then bound to the A-site on the small ribosomal subunit by elongation factor Tu. The ribosome then catalyzes the formation of a peptide bond between the tRNA-bound amino acid and the growing protein chain. Elongation factor G then moves the peptidyl-tRNA complex to the P-site of the ribosome. The tRNA-peptide link is cleaved on arrival of the next tRNA at the A site. The uncharged tRNA then moves to the E site and is released. Specific tRNAs are usually abbreviated with the 3-letter amino acid designation in subscript. Thus the tRNAs for alanine are abbreviated tRNAala. See also, aminoacyl-tRNA.
V-ATPase This is the eukaryotic version of the F-ATPase, q.v. V-ATPase is constructed slightly differently so that the "forward" reaction is favored. That is, it functions as an ATP-dependent ion pump. It is used, for example, to pump protons into digestive vacuoles to maintain very low pH inside the vacuole. V-ATPases are also found in various Archaea and in some Eubacteria which seem to have acquired the archaean species by lateral gene transfer. The archaean version is sometimes referred to as "A-ATPase." Finbow & Harrison (1997)
ATW??????, page uploaded RFVS111017, edited RFVS111214
Text placed in the public domain