| Demospongiae | ||
| Porifera | Demospongiae - 4 |
| Metazoa | Metazoa | Metazoa | ||||
| Time |
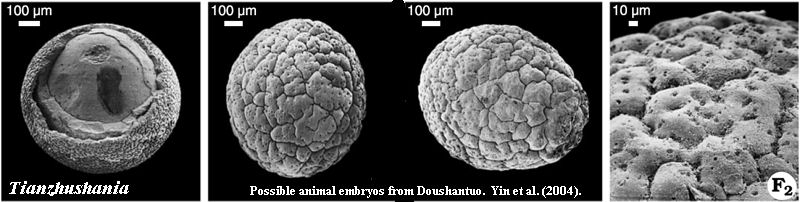
There is little doubt that some kind of sponges go back to the Early Ediacaran. Li et al.(1998). However, as Hagadorn et al. 2006) have noted, the (probable) embryos of the Ediacaran Doushantuo Formation may not be demosponges, since there is nothing resembling a parenchymella and little evidence of epithelium-like tissues. However, there is nothing which requires that these embryos have any relationship to the adult sponges of the Doushantuo. The adult sponges from Doushantuo are mainly globular, "but a few are tubular ... and range in size from 150 to 750 µ. Li et al. (1998). Only thin, monaxonal spicules are present, and these are randomly located and oriented. The spicules are between 0.5 and 1.0 µ in diameter, but as much as 60 µ long. The very largest are 4 µ in diameter and 100 µ long. Id. Li et al. believe these are demosponges based on the presence of silica and well-defined spongeocoels.
 By the Terreneuvian, tubular demosponges, such as Leptomitus, Leptomitella, and Paraleptomitella dominated the spiculate sponge fauna, together with spiny, funnel-shaped sponges such as Choiaella. Hagadorn (2002). Some of these were persistent, widely dispersed forms. The pictured example of Leptomitus comes from the Middle Cambrian of Spain, and similar examples are known from the Burgess Shale of early Middle Cambrian Canada. García-Bellido Capdevilla (2003).
By the Terreneuvian, tubular demosponges, such as Leptomitus, Leptomitella, and Paraleptomitella dominated the spiculate sponge fauna, together with spiny, funnel-shaped sponges such as Choiaella. Hagadorn (2002). Some of these were persistent, widely dispersed forms. The pictured example of Leptomitus comes from the Middle Cambrian of Spain, and similar examples are known from the Burgess Shale of early Middle Cambrian Canada. García-Bellido Capdevilla (2003).
 As Dornbos et al. 2005) point out, many of these sponges were still essentially adapted to a firm Neoproterozoic-style substrate, held together as a bacterial mat. However, as the Cambrian progressed, this mat was looking increasingly moth-eaten and patchy, chewed up by "worms" and replaced by a soupy mud that left little purchase for attachment. As we've mentioned in connection with other early sponge groups, one of the key adaptations required for survival in these increasingly sloppy environments of the Middle and Furongian was the ability either to grow large quickly, or to grow attached to someone else who had already worked out a practical method for living in mud. As often as not, that “someone” was a demosponge, at least until the rise of stromatoporoids in the Ordovician. Accordingly, from the Furongian into the Silurian, demosponges literally supported a biota of bryozoans, unidentifiable worms, rugose corals, and articulate brachiopods. Taylor & Wilson (2003).
As Dornbos et al. 2005) point out, many of these sponges were still essentially adapted to a firm Neoproterozoic-style substrate, held together as a bacterial mat. However, as the Cambrian progressed, this mat was looking increasingly moth-eaten and patchy, chewed up by "worms" and replaced by a soupy mud that left little purchase for attachment. As we've mentioned in connection with other early sponge groups, one of the key adaptations required for survival in these increasingly sloppy environments of the Middle and Furongian was the ability either to grow large quickly, or to grow attached to someone else who had already worked out a practical method for living in mud. As often as not, that “someone” was a demosponge, at least until the rise of stromatoporoids in the Ordovician. Accordingly, from the Furongian into the Silurian, demosponges literally supported a biota of bryozoans, unidentifiable worms, rugose corals, and articulate brachiopods. Taylor & Wilson (2003).
After stromatoporoids became a major factor, demosponges tended to adopt an encrusting habit and themselves become epibionts. With the extinction of Paleozoic stromatoporoids at the end of the Devonian, demosponges again became important as substrates for Carboniferous and Permian marine fauna. Id. During the Mesozoic, and particularly the Jurassic, demosponges evolved a large number if different types: new encrusters, massive lithistid forms, and perhaps also the Mesozoic stromatoporoid groups. They remain the dominant class of sponges today, and have even invaded fresh water habitats during the Cenozoic..
All phylogenetic analyses using the basic morphological characters results in an animal tree with monophyletic sponges. Sperling et al. 2006). On the other hand, studies using molecular sequences have yielded just about every conceivable result except monophyly. Id. For a variety of reasons, we place relatively little faith in either approach. In some ways, the case against molecules is easier to make. We discussed it elsewhere, and nothing has happened in the last two years which signals any change in the foundations of statistics. Sequence phylogenies are not capable, even in theory, of sorting out lineages which diverged in a short period of time and the problem becomes worse when the lineage splitting events happened long ago. It isn't always a question of getting more DNA, more species, or improved calculations. We may be up against fundamental mathematical limitations. Rokas et al. (2003); Rokas et al. 2005). Worse, unless the fossil record is extremely good, we won't even know when we have crossed the border between low resolution and white noise. However, the continued failure of sequence phylogenies to yield consistent results is a strong indicator that we are over the frontier with the sponges.
At the same time, parsimony techniques using morphological characters are also likely to be futile. The lithistid sponge pictured above has so few characters in common with a fruit fly or zebra fish, that we have little basis for morphological comparison. Even where we can identify morphological similarities, it is rarely possible to tell homology from coincidence with any confidence. So how can we parse the evolution of demosponge, Drosophila, and Danio?
Here, as in many other places, we have tried to suggest a middle way. "Suggest" is perhaps a poor choice of words, since it implies that the idea was ours in the first place. It isn't. It is an approach which workers as diverse as Tom Cavalier-Smith and Sean Carroll have been converging on for over a decade. At the 2007 SVP meeting, several graduate students I spoke with seemed headed in the same direction. Molecular sequences have a huge number of problems. Their real advantage is funding, and the fact that they don't require much knowledge of biology or evolution. But organisms don't live and die based on molecular sequence. The key in biochemistry, as in anatomy, is structure. We may be able to predict structure from sequence, but that doesn't help us if we continue to attempt to compare sequences instead of structures. While gross morphology changes too fast to compare organisms which are distantly related, or diverged long ago, the structure of their molecules, the morphology of cellular components, and various biochemical patterns of development, are more stable. What's more, the characters are frequently objective, reproducible, and involve unquestionable homology.
We have deliberately refrained from giving this approach a catchy name. That's a job for others who have earned the right to make names stick. We can only hope that they do not work at Harvard. Here we've tried to look at a few of the appropriate characters: spicule formation, collagen structure, ciliary rootlets, and a bit of embryology. Unlike sequence analysis, this stuff requires lots of biology. As sponge amateurs, our ability to analyze the facts is limited. It seems to fit together in a reasonable way, which has encouraged us to crawl far out on an unorthodox phylogenetic limb. With luck, that limb will begin to bend under the burden of weightier opinions. ATW071125.
ATW071122