
| Palaeos: |  |
Gruimorpha |
| The Vertebrates | Falconiformes & Allies |
| Page Back | Unit Home | Unit Dendrogram | Unit References | Taxon Index | Page Next |
| Unit Back | Vertebrates Home | Vertebrate Dendrograms | Vertebrate References | Glossary | Unit Next |
|
Abbreviated Dendrogram
AVES |--GALLOANSERAE Gruimorpha |--+--Gruiformes | `--Podicipediformes `--+--+--Strigiformes | `--+--+--Caprimulgiformes | | `--Coliiformes | `--Apodiformes `--+--Musophagidae `--+--+--Falconiformes | `--Cuculiformes | |--Cuculidae | `--Neomorphidae `--+--Piciformes | PASSERIFORMES |
Contents
|
Chronologically, this is the second attempt in these Notes to examine the phylogenetic position of one of the major bird orders. The first (see Anseriformes) degenerated quickly into impasse between the major contending viewpoints. Here we will, instead, take a firm position, and a very heterodox one at that, based on some largely ignored (even by the authors) findings in molecular phylogenetics from Mindell et al. (1998).
As this site is oriented to organismal biology, a little explanation or reminder is in order. Phylogenetic systematics attempts to apply mathematical techniques, and occasionally logic, to tease phylogeny out of physical characteristics. Molecular phylogenetics uses similar techniques to derive phylogeny out of biomolecules. Unfortunately, and quite possibly wrongly, current work has focused on primary structure. That is, systematists have attempted to treat the sequences of DNA bases or the amino acid sequences of proteins as if they were strings of characters. It is certainly appealing to look at genes and proteins in this fashion. Each base or amino acid is a discrete item of information, and -- unlike many organismal characters -- there is usually little dispute about what the sequence is and no intermediate states exist. DNA has only four bases, after all, and there are only twenty-odd naturally occurring amino acids. One merely compares homologous sequences in different species and calculates the most parsimonious path from some ancestral form. What could be simpler?
 As it turns out, it is not simple at all. There are a large number of reasons
for this difficulty, only a few of which will be discussed here. One problem
which has not received enough attention, or any at all, is the naive reliance on
primary structure itself. "Primary" structure is the bare sequence of
nucleotides or amino acids. "Secondary" structure relates to simple,
stable structural units normally cemented by hydrogen bonds: double-stranded
regions of nucleic acids, alpha helices or beta sheets in proteins.
"Tertiary" structure is the labile, full three-dimensional structure
of the functional molecule. "Quaternary" structure refers to the
temporal and chemical association of the functional molecule with cofactors,
substrates and other subunits of more complex aggregations.
As it turns out, it is not simple at all. There are a large number of reasons
for this difficulty, only a few of which will be discussed here. One problem
which has not received enough attention, or any at all, is the naive reliance on
primary structure itself. "Primary" structure is the bare sequence of
nucleotides or amino acids. "Secondary" structure relates to simple,
stable structural units normally cemented by hydrogen bonds: double-stranded
regions of nucleic acids, alpha helices or beta sheets in proteins.
"Tertiary" structure is the labile, full three-dimensional structure
of the functional molecule. "Quaternary" structure refers to the
temporal and chemical association of the functional molecule with cofactors,
substrates and other subunits of more complex aggregations.
Ultimately, proteins function as very complex three dimensional structures. Similarly, many non-coding sections of nucleic acids function as three-dimensional structures specific for the (quaternary) binding of regulatory molecules, transport proteins, polymerases and their regulators, and so on. The change of a single nucleotides may thus have little or no effect on functionality in one position, but, in another position, it may block the binding of an important regulator or cause the ultimate gene product (i.e. the protein the gene codes for) to behave in unexpected ways. In many cases, it would actually make more sense if molecular phylogenetics were carried out more like normal cladistics, using descriptive attributes of the shape, surface characteristics, and articulations of the gene product, rather than the primary structure of the gene. As matters stand, we are not only comparing apples and oranges, but also have no idea at all which is an apple and which is an orange, grapefruit, or wild mountain boysenberry.
 Certainly different portions of the same protein evolve at wildly different
rates. Recent work has also shown that the genome is a much more lively place
than originally believed. Genes and pieces of genes are moved, flipped, broken
up, used in multiple places, and reorganized with remarkable frequency.
Consequently, homology is often more difficult to determine in molecules than in
whole organisms. The net result is that using statistical analysis on raw primary
sequence data sometimes leads to rather unlikely results.
Certainly different portions of the same protein evolve at wildly different
rates. Recent work has also shown that the genome is a much more lively place
than originally believed. Genes and pieces of genes are moved, flipped, broken
up, used in multiple places, and reorganized with remarkable frequency.
Consequently, homology is often more difficult to determine in molecules than in
whole organisms. The net result is that using statistical analysis on raw primary
sequence data sometimes leads to rather unlikely results.
A rather clear example of this is the massive study of Mindell et al. (1997). David Mindell of the University of Michigan is one of several workers attempting to determine the ordinal relationships of birds using molecular methods. He has worked mainly with mitochondrial genes. Mitochondria are believed to have evolved very early as bacterial endosymbionts, independent cells living within other cells. They are found as common organelles within virtually all cells of all eukaryotes and are the principle site of oxidative metabolism. They retain their own DNA genome ("mtDNA") which does not normally recombine and passes solely through the maternal line. The mitochondrial genome is quite small, but contains, for example, genes for unique mitochondrial ribosomal RNAs, transfer RNAs and certain enzymes involved in oxidative metabolism.
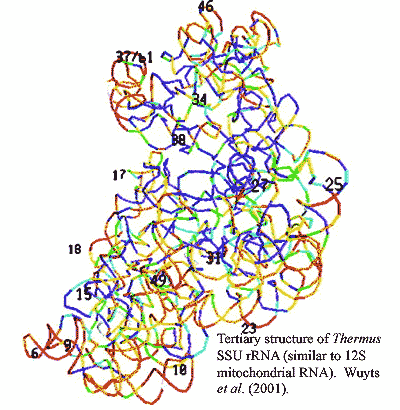
Since there are a great many mitochondria per cell, all with the same DNA complement, this appears to be an experimentally easy system to work with for molecular evolutionary studies. Unfortunately, it really isn't. Mindell worked, in part, with the 12S rRNA gene. 12S rRNA is a structural RNA that has an incredibly elaborate secondary structure, with numerous loops, turns, and double-stranded regions. The sequences involved are often highly conserved, but they are also highly correlated. Other sections are hypervariable -- not only in sequence, but in length, so that determinations of homology become impossible. As to tertiary structure -- the image from Wuyts et al. (2001) will give you some idea of the problem. Ultimately, Mindell was forced to eliminate substantial portions of the gene from consideration. (The double stranded regions may also interfere with the polymerase chain reaction used to amplify DNA for sequencing and yield aberrant results.)
In the end, after what must have been a huge effort, Mindell was left with an essentially untenable phylogeny which may be diagrammed as follows:
Root (Aythya, Cygnus)
|--Passeriformes (7/7)
`--+--+--+--Ciconiiformes (1/2)
| | `--+--Falconiformes (1/13)
| | `--Strigiformes (1/12)
| `--+--Trogoniformes (1/1)
| `--Strigiformes (11/12)
`--+--Falconiformes (11/13)
`--+--+--Pelicaniformes (1/1)
| `--Cuculiformes (1/3) cuckoo
`--+--Gruiformes (1/2) Turnix
`--+--+--+--+--Ciconiiformes (1/3)
| | | `--Coraciiformes (1/1)
| | `--Apodiformes (1/1)
| `--+--+--+--Gruiformes (1/2)
| | | `--Cuculiformes (1/3) Turaco (Musophagidae?)
| | `--Procellariiformes (1/1)
| `--Charadriiformes (2/2)
`--+--+--Falconiformes (1/13) Sagitariid
| `--Caprimulgiformes (1/1)
`--+--Phoenicopteriformes (1/1)
`--Cuculifomes (1/3) hoatzin
The figures in parentheses refer to the number of species at that position compared to the total number presumptively belonging in that order. That is "Falconiformes (1/13)" means that one of the 13 traditionally falconiform species in the study was found at that position.
There is no sense in beating this dead horse. The hoatzin simply isn't the closest living relative of the flamingo; nor are they together the sister group of the secretary bird. The cuckoo is not related closely to the pelican. Other, more detailed sections of the paper, largely using the cytochrome oxidase I gene, were useful and informative. This part, for whatever reason, was simply a bust.
Mindell has since moved on. His more recent projects involve tracking bird phylogeny by detecting the presence of avian virus signals in the nuclear genome. This appears to be a singularly inventive and probably productive line of work. However, in the interim, he and his co-workers had one more shot at the mitochondrial approach. Mindell et al (1998). In this paper, Mindell noted that the general vertebrate gene order on the mitochondrial chromosome (call it "A") is well-known. More recently, it was discovered that bird mitochondria display a different gene order ("B"). Mindell discovered that certain bird orders have yet a third arrangement ("C"). C appears to be derived from B by a tandem repeat. That is, a section of mtDNA has been repeated, and the repeat is inserted immediately after the "original" on the mitochondrial chromosome. The original duplication event(s) were followed by partial deletion of the non-coding and non-functional duplicated control region. See also, Bensch & Härlid (2000). Mindell found that sequence C occurred in the falconiforms, cuculiforms, piciforms and in suboscine passeriforms. Unfortunately, when mapped against certain widely used avian ordinal phylogenies, the distribution of C was all over the map. Mindell concludes that C had "multiple independent origins" within Neornithes.
 It is distinctly possible that Mindell was too quick to discard his result.
Mapped against the "consensus" phylogeny used in these Notes, the
distribution of C is closely clustered in the advanced gruimorphs, allowing for
one reversal in the oscine passeriforms. In fact, the clustering was so striking
that the phylogeny here has been re-arranged on the assumption that this is in
fact the true phylogenetic signal. This conformation does require that the
cuculiforms be separated from the Musophagidae. This is an unhappy result
because the hoazin (Opisthocomus hoatzin) seems to be close to both
cuculiforms and musophagids. Hughes
& Baker (1999); Hedges et al.
(1995). However, the separation need not be large, and the final tree is
otherwise fairly reasonable.
It is distinctly possible that Mindell was too quick to discard his result.
Mapped against the "consensus" phylogeny used in these Notes, the
distribution of C is closely clustered in the advanced gruimorphs, allowing for
one reversal in the oscine passeriforms. In fact, the clustering was so striking
that the phylogeny here has been re-arranged on the assumption that this is in
fact the true phylogenetic signal. This conformation does require that the
cuculiforms be separated from the Musophagidae. This is an unhappy result
because the hoazin (Opisthocomus hoatzin) seems to be close to both
cuculiforms and musophagids. Hughes
& Baker (1999); Hedges et al.
(1995). However, the separation need not be large, and the final tree is
otherwise fairly reasonable.
Bensch & Härlid argue that certain internal sequence homologies demonstrate multiple convergent origins for this rearrangement. However, their results are unconvincing and, at least to my eye, tend to support the opposite and more parsimonious explanation: a single duplication event followed by a single reversal in the passeriforms. Briefly, the Swedish group posits that, if the duplication occurred once, the duplicated sequence in all families would look like some hypothetical ancestral sequence. If duplication occurred many times after the radiation of the neornithine orders, each of the duplicated sequences should be closer to the homologous sequence for the particular family in which it is found.
The hypothesis itself is far from airtight. Since the duplicated sequence is not functional and is subject to completely random deletions and mutations, it may be impossible to locate true homologies amid the noise. This turns out to be exactly what is observed. The "homologies" claimed by the Swedish group are a few, scattered, short stretches of a dozen nucleotides or less, only a few positions of which are phylogenetically significant. The correspondence between this genetic static and the unknown sequence from an unknown ornithurine ancestor is impossible to determine or compare.
Recently (shortly after the original version of this essay, in fact!), Macey et al. (2000) similarly criticized Mindell's interpretation of his results for much the same reasons. Macey's group found that alterations in mitochondrial genome structure are extremely stable and are strong phylogenetic markers in a group of acrodont lizards. Since lizards can be sorted out far better than birds, using more standard phylogenetic methods, the lizard system is a much better test of the stability of mtDNA rearrangements. The alteration in acrodont mtDNA found by Macey is a simple reversal in the order of two mitochondrial tRNA-coding genes, both of which remain functional. Thus both copies are subject to stabilizing selection, and would be expected to diverge in a more gradual and traceable manner. Thus, this is a far better system than the interordinal relationships of birds from which to draw conclusions about stability. If Macy is correct, and mtDNA rearrangements are very rare and very stable, it may be concluded that the neornithine rearrangements most probably occurred only once. Thus, Mindell may have found, but discarded, a critical part the true tree which has eluded avian taxonomy for so long. ATW 000904 revised 051126.
 We recently discovered a new site -- new to us, anyway -- the Searchable
Ornithological Research Archive (SORA)
from the University of New Mexico at Albuquerque. SORA contains a number
of ornithological journals complete to about 2000. This includes Auk
back to 1884, Condor to 1899, as well as some less well-known or
discontinued journals. Ornithology is not a field in which everything more
than two years old is obsolete. Consequently, there's a lot of useful
information here for the asking. We took it for a test drive on the
Cuculidae.
We recently discovered a new site -- new to us, anyway -- the Searchable
Ornithological Research Archive (SORA)
from the University of New Mexico at Albuquerque. SORA contains a number
of ornithological journals complete to about 2000. This includes Auk
back to 1884, Condor to 1899, as well as some less well-known or
discontinued journals. Ornithology is not a field in which everything more
than two years old is obsolete. Consequently, there's a lot of useful
information here for the asking. We took it for a test drive on the
Cuculidae.
The site has a full-text search feature, which ought to give the same results as a Google site search. For some reason, the full-text search actually generates 50% more results (149 hits) than Google (101 hits). Possibly, Google's robots don't travel to SORA often enough to keep up. SORA uses a very promising, and very fast, new open-source indexing system called Swish-e. Swish-e may simply be more thorough than Google for pdf file collections, an area in which the desk-top version of Google is somewhat weak (judging from personal experience). The subject index option only searches article titles. Unless you're looking for something quite specific, it's not terribly useful. By default, the full-text search lists by relevance; so this will usually be the better option. A minor warning: the search engine appears to give you a choice of searching full text or by field. However, it searches by both. If you left a name in the author field when turning to full text, you will only recover papers which satisfy both criteria, no matter which search mode you select.
Just to get a flavor of the archive, we looked at the first few papers listed for Cuculidae. The first is Berger (1960). Andrew Berger was a well-known avian anatomist who published extensively during the 1950's and 60's. Much of his anatomical work was focused on the Cuculidae. As this paper shows, Berger was willing pursue detailed anatomy -- particularly myology -- to an extent that few academics would even consider today; and, even if they did, no one would fund it. Out of interest, we ran some of the muscles Berger discusses back through the SORA search engine. In many cases, the only papers which mention these details are other works by Berger. Berger's nomenclature seems to be standard (he literally wrote the book on avian myology), so we can only conclude that this kind of work is essentially a lost art. Berger not only describes these details, but discusses them on a comparative basis, with numerous comments on their relationship to locomotion, phylogeny, and mode of life. In short this is useful information which is not available elsewhere, and is unlikely to become available in the forseeable future.
The second paper listed is Hughes (1996), a much more recent study. Five years ago, we wrote a short piece on Slikas (1998) and the utility of behavioral characters for avian phylogenetics. We wondered, at the time, if anyone else was using that methodology. Prof. Hughes, now at Lakehead University in Ontario, published the cited work as a graduate student, as a run-up to a better known osteological analysis she published later [1]. However, only the 1996 paper combines osteological, behavioral and ecological characters. It is particularly interesting to compare Hughes' conclusions with Berger (1960). Berger refuses to commit to any particular phylogeny, but he drops enough hints -- mostly by way of rebuttal to others -- to constrain his phylogeny. The two trees have some striking similarities, despite the differences in approach.
So, for example, Hughes and (probably) Berger place Tapera with the obligate brood parasites of the Cuculidae, rather than in its classical position among the Neomorphidae.
[1] Hughes, JM (2000), Monophyly and phylogeny of cockoos (Aves, Cuculidae) inferred from the osteological characters. Zool. J. Linn. Soc. 130: 263-307. We have not personally reviewed this work.
 Falconiformes:
falcons, hawks, eagles, Old World vultures.
Falconiformes:
falcons, hawks, eagles, Old World vultures.
Range: from the Middle Eocene
Phylogeny: Gruimorpha ::::: Cuculiformes + *.
Characters: head fairly large; sharply hooked beak with cere (soft mass) on proximodorsal surface housing nostrils; wings long and fairly broad (soaring) with outer 4-6 primaries emarginated; strong legs and feet with raptorial claws and opposable hind claw; strong flyers; almost all carnivorous; diurnal or crepescular; exceptionally long-lived, with low reproductive rate; long, very fast-growing fledgling stage, followed by 3-8 weeks of nest care after first flight, and 1-3 years as sexually immature; sexes have conspicuously different sizes; monogamy common.
Links: kingdom; animal diversity falconiformes; Untitled; Íslenskir ránfuglar (Icelandic?); Raptor Rehabilitation of Ky. - Kentucky Raptor Species; Related Links; Bird Sounds Digitally Recorded; Falconiformes Links; BIRDNET: Falconiformes; Orden Falconiformes; Raptor Index; Falconiformes of the World: A Checklist; The birds of prey site of Robert Goedegebuur; Falconiformes; Falconiformes; Google Directory - Falconiformes; Birds of Prey - Order Falconiformes; <br><b> Order Falconiformes: Hawks & Eagles; Falconiformes (Spanish); Aves: Falconiformes - Falkenartige (German); Natural Foods of Falconiformes & Strigiformes: Falcons & Owls; Otrya Sokoloobrazhnye (Russian); Falconiformes; Falconiformes index (Japanese & English).
Image: "Accipiter" from the Aberdeen Bestiary (XIIth Century?) ATW020704.
Cuculiformes: cuckoos, roadrunners, possibly hoatzin.
Range: from the Late Eocene.
Phylogeny: Gruimorpha ::::: Falconiformes + * : Cuculidae + Neomorphidae.
Characters: Small to moderately large (15-90 cm) forest insectivores with a marked tendency to brood parasitism, i.e. the habit of laying eggs in the nests of other species. Beak moderate sized, often with slight down-curve; slender body; long tail; moderately long , eutaxic wings; legs often long; feet zygodactylous or facultatively zygodactyl; many species are brood parasites, or have communal nests; brood parasites mimic egg coloration of host (specific in the female line); typically insectivores, but some hunt small vertebrates or eat fruit; fairly vocal, with most vocalizations by male; long, unguided migrations by yearlings known. Cuculiforms are sometimes placed closer to Psittaciformes.
Links: Order Cuculiformes; Dierentuin.Net Dieren Database Koekoeken; Cuckoo - Enchanted Learning; FLMNH bird photos archive page 22; Australian Cuckoos; BIRDNET: Cuculiformes species list; Aves: Cuculiformes; World Birds Taxonomic List:.
References: Bensch & Härlid (2000); Hedges et al. (1995); Hughes & Baker (1999); Macey et al. (2000); Mindell et al. (1997); Mindell et al. (1998). ATW010622.
 Cuculidae:
Coccyzus, cuckoos? "Cuculidae" is sometimes used to
refer to 3 different groups: (a) Cuculiformes; (b) Cuculiformes except
Neomorphidae; and (c) the Old World cuckoos, or Cuculinae. At the moment, it is
used here in the second, paraphyletic, sense as a placeholder until there is an
opportunity to explore the group further.
Cuculidae:
Coccyzus, cuckoos? "Cuculidae" is sometimes used to
refer to 3 different groups: (a) Cuculiformes; (b) Cuculiformes except
Neomorphidae; and (c) the Old World cuckoos, or Cuculinae. At the moment, it is
used here in the second, paraphyletic, sense as a placeholder until there is an
opportunity to explore the group further.
Phylogeny: Cuculiformes : Neomorphidae + *.
Characters: strong legs & feet (some forms terrestrial); tails long; largely insectivorous; frequent brood parasites;
Links: Cuckoo family (Best on the Web); Aves: Cuculiformes; Bird families; Cuculidae Family; .
Image: Coccyzus americanus by Dr. Dan Sudia, courtesy of University of Georgia Museum of Natural History and reproduced under standard terms. ATW000902.
 Neomorphidae:
roadrunners & ground cuckoos: Dromococcyx, Geococcyx, Morococcyx,
Neomorphus
Neomorphidae:
roadrunners & ground cuckoos: Dromococcyx, Geococcyx, Morococcyx,
Neomorphus
Phylogeny: Cuculiformes : Cuculidae + *.
Characters: tails long; legs strong; feet zygodactyl.
Links: Bip-Bip; Roadrunner; Bird Data Project - Search Results (diversity data); AVIFAUNA - Picchio verde . . . l'altro web site - Birds- DNA ... (distribution & biomes); Cuckoo family; Roadrunner - The Bird and Cartoon; mbev-20-09-03 1484..1499 (molecular phylogeny). ATW040104
 Piciformes:
woodpeckers, toucans.
Piciformes:
woodpeckers, toucans.
Range: from the Late Eocene? middle Miocene?
Phylogeny: Gruimorpha ::::: Passeriformes + *.
Characters: skull thick; tongues long and barbed or sticky; hyoid may be highly modified for protrusion of tongue; tail feathers stiffened for support in climbing or perching; feet and legs modified for vertical climbing; zygodactyl feet or three-toed (loss of I); largely insectivorous, some fruit; almost all excavate nests in trees, earth, etc.; a few are brood parasites (honey guides); multiple songs; tendency to be aggressive and territorial.
Links: Red-Brested Toucan Skull Replica offered by Skulls Unlimited; Order Piciformes; Aves: Piciformes - Spechte; Dierentuin.Net Dieren Database Spechten en verwanten; FLMNH bird photos archive page 27; World Birds Taxonomic List: Genera and species with citations.; Patuxent Bird Identification InfoCenter.
| Page Back | Unit Home | Page Top | Page Next |
Checked ATW050730