Eubacteria
 This is the Domain of the Germs. The Eubacteria, in their hundreds of trillions, are the reasons you weren't allowed to pick up the candy you dropped on the floor or eat that egg salad that looked so good a week ago. They are behind every torture you have ever suffered at the hands of the dentist, and they are the root cause of childhood afflictions from antiseptics to acne. They have a lot to answer for. Then again, they're probably not too happy with us, either. Its hard to tell, since bacteria don't go in much for light conversation or email rants. They grow, or they don't, just as they have for the last three or four billion years, and without making much fuss about it. Bacterial psychology is thus a rather limited field. But such simplicity has its advantages. We are never tempted to paint human thoughts and emotions onto a 4 µ long pill-shaped blob of protoplasm. We can safely view the bacterium for what it is, a small biochemical machine, without having to steer the usual narrow passage between the twin perils of anthropomorphism and reductionism.
This is the Domain of the Germs. The Eubacteria, in their hundreds of trillions, are the reasons you weren't allowed to pick up the candy you dropped on the floor or eat that egg salad that looked so good a week ago. They are behind every torture you have ever suffered at the hands of the dentist, and they are the root cause of childhood afflictions from antiseptics to acne. They have a lot to answer for. Then again, they're probably not too happy with us, either. Its hard to tell, since bacteria don't go in much for light conversation or email rants. They grow, or they don't, just as they have for the last three or four billion years, and without making much fuss about it. Bacterial psychology is thus a rather limited field. But such simplicity has its advantages. We are never tempted to paint human thoughts and emotions onto a 4 µ long pill-shaped blob of protoplasm. We can safely view the bacterium for what it is, a small biochemical machine, without having to steer the usual narrow passage between the twin perils of anthropomorphism and reductionism.
However, to say that the Eubacteria are biochemical machines is not to belittle them. Consider Gomphosphaeria Kützing 1836, the somewhat larger than average, but otherwise undistinguished, phytoplanktonic cyanobacterium [1] on the right. If we allowed a single cell of Gomphosphaeria to grow and divide under optimum conditions for only about 4.5 days we would be up to our armpits in Gomphosphaeria over the entire surface of the Earth [2]. Even man's most perfect machines, for example the 1976 Toyota Corolla, couldn't come close to matching this kind of performance.
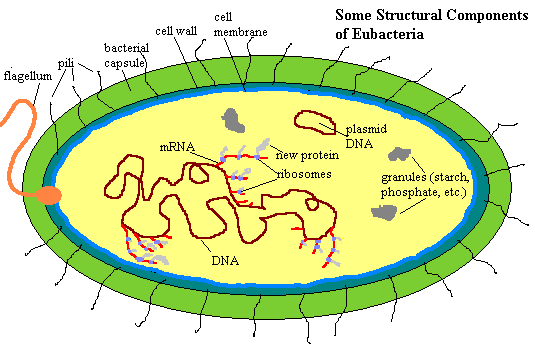 This page will be devoted to considering the basic structural and functional units of the Eubacteria as biological machines. More specific matters, as well as considerations of ecology, phylogeny and evolution will be taken up in connection with more the specific groups to which these matters pertain. So, how are these machines put together? Some of the basic parts are shown in the figure on the left. Look at it carefully, because we have a good bit to cover, starting from outside.
This page will be devoted to considering the basic structural and functional units of the Eubacteria as biological machines. More specific matters, as well as considerations of ecology, phylogeny and evolution will be taken up in connection with more the specific groups to which these matters pertain. So, how are these machines put together? Some of the basic parts are shown in the figure on the left. Look at it carefully, because we have a good bit to cover, starting from outside.
This survey also takes us into some rather dense discussion of cell biology, a little biochemistry, and even a smidgen of thermodynamics. In fact, this page will serve, for the present, as a very condensed introduction to cell biology for the entire site. If you don't want to hear about that stuff, you probably shouldn't be reading about bacteria—or single-celled organisms of any kind.
Geometry and mechanics dictate that the first structures we encounter in any cell—and most multicellular organisms, for that matter—are structures dealing with movement, sensation, and interaction with the world outside. In our model eubacterium, these include a flagellum and a system of pili.
The bacterial flagellum looks a bit like the eukaryotic organelle of the same name, but it is an entirely different structure. The "tail" portion has no microtubules and consists more or less of an extended filament of a single protein, flagellin. The tail is anchored on what amounts to a rotor. The rotor extends through the outer layers of the cell into the cytoplasm. The system actually works something like a propeller, with the rotor forcing the flagellum to turn in a spiral. The motive force is supplied by sodium or hydrogen ions flowing down a concentration gradient from the outside.
This ion gradient system is the same basic mechanism which is used in a number of other well-known systems, for example in some of the "light reactions" of photosynthesis. It is worth knowing reasonably well. The usual currency of energy in the cell is adenosine triphosphate ATP). When the cell has ATP to spare, it uses ATP to pump certain ions here, sodium) out of the cell. Since the sodium concentration is higher outside the cell, the pumps have to pump against the gradient using the energy of ATP. This makes the entire cell a sort of storage battery. To use that stored energy, the cell merely allows some of that sodium to flow back in by means of specialized sodium channels. These channels span the cell membrane and cell wall. When the sodium channel proteins are activated by signals inside the cell and come in contact with a sodium ion outside the cell, they change shape, allowing the ion into the cell and, at the same time, performing some useful work, such as turning the "rotor" of a flagellum [3]. The key concept is that the ion channel system takes small bits of energy (ATP molecules) which are all the same and are dispersed throughout the cell, and ultimately concentrates the energy for use at a time and place controlled by specific "signal" molecules that open specific ion channels linked to specific mechanical tasks—a very elegant system!
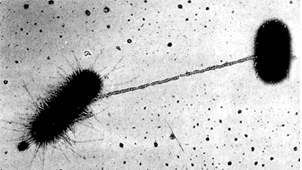 The bacterial pili are the bug's equivalent of hands, both as tactile, sensory structures and as tools for grabbing onto and manipulating things at a distance. Our diagram is a bit misleading, since the pili can be quite long. See image (~20,000X) at right from Le revêtement cellulaire des cellules procaryotes. As this image shows, the pilus can serve as a guide for the formation of a cytoplasmic bridge, as for the exchange of DNA. This is a rare, but important event with profound implications for bacterial evolution. Bacteria are not terribly fastidious about who they exchange DNA with. Thus genes can be acquired from unrelated bacteria, and even from non-bacteria. For example, DNA is being exchanged between Escherrichia and a virus in the image. For this reason, "lateral inheritance" of genes from unrelated organisms is quite frequently observed in bacteria.
The bacterial pili are the bug's equivalent of hands, both as tactile, sensory structures and as tools for grabbing onto and manipulating things at a distance. Our diagram is a bit misleading, since the pili can be quite long. See image (~20,000X) at right from Le revêtement cellulaire des cellules procaryotes. As this image shows, the pilus can serve as a guide for the formation of a cytoplasmic bridge, as for the exchange of DNA. This is a rare, but important event with profound implications for bacterial evolution. Bacteria are not terribly fastidious about who they exchange DNA with. Thus genes can be acquired from unrelated bacteria, and even from non-bacteria. For example, DNA is being exchanged between Escherrichia and a virus in the image. For this reason, "lateral inheritance" of genes from unrelated organisms is quite frequently observed in bacteria.
Pili perform a great many functions, and consequently are structurally quite diverse. Typically the backbone of the pilus is made up of a long chain protein or polysaccharide (sugar chain) with some type of functionally specific arrangement at the tip. One function of considerable clinical interest is cell-to-cell recognition. The complex array of carbohydrates in and on the pili are the method by which bacteria recognize other cells, and are recognized by them. So, for example, one strain of a germ may be harmless to us while another, differing only in a few sites, may be a deadly pathogen if it recognizes our cells as food, or has surface features which our cells do not recognize as dangerous. Shorter pili, usually referred to as fimbriae, are a structurally distinct group of extrusions which operate mostly in bacterial attachment to substrate or to other cells.
The next structure we may encounter is the bacterial capsule. The composition, nature, and even existence of the capsule are highly variable, even within a species. It may be composed of polysaccharide or protein, may be tightly or loosely bound, or it may not be found at all. The capsule may be best thought of as an extremely dense layer of short pili. It functions in cell attachment, resistance to desiccation, and as a defense to being swallowed (phagocytosed) by other cells. Streptococcus pneumoniae is a case in point. The R strain of Streptococcus has no capsule and will not cause disease. It is readily engulfed by human cells. The bacterium is then encased within the membranes of a "vacuole" (a membrane-bound bubble inside the cell) and digested by lysozymes within the vacuole. The S strain has a capsule which prevents digestion. It uses the normally lethal environment of the phagocytic vacuole to grow, replicate and prepare to devour the cell from inside.
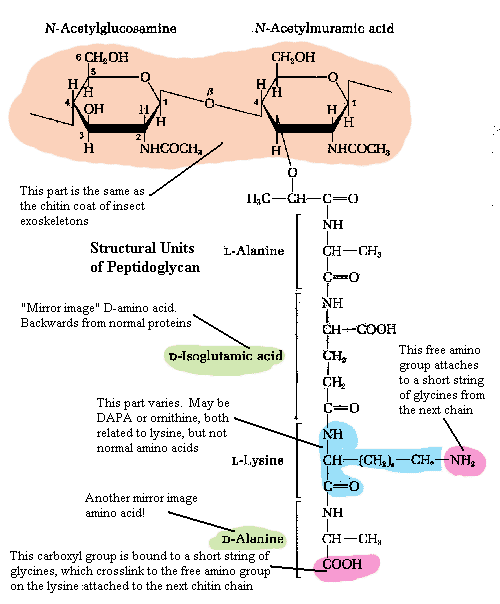 Within the capsule is the relatively rigid cell wall[4]. The fundamental scaffolding of the cell wall is peptidoglycan. See, generally, The Cell Wall. Peptidoglycan is an absolutely bizarre material. As this is not a biochemical essay, we will have to skip over much of the good stuff.
Within the capsule is the relatively rigid cell wall[4]. The fundamental scaffolding of the cell wall is peptidoglycan. See, generally, The Cell Wall. Peptidoglycan is an absolutely bizarre material. As this is not a biochemical essay, we will have to skip over much of the good stuff.
The first element of peptidoglycan is a chain of repeating sugar molecules (a slightly modified glucose, N-acetylglucosamine). This part of the structure is precisely the same as chitin, the material which makes up the exoskeleton of insects and, in more or less modified form, in almost all arthropods. Significantly, it is also found in the radular "teeth" of molluscs, the setae (bristles) and jaws of annelid worms, and the cell walls of Fungi. So, this is exceedingly ancient stuff, possibly predating the split between bacteria and metazoans.
However, in the Eubacteria, every second sugar residue is linked at the 3-position with an amino acid, threonine, which in turn leads on to a strange and unique chain of amino acids (i.e., a peptide chain). The ordinary amino acids which make up all proteins are asymmetrical. That is, they can occur in left-handed (L) or right -handed (D) forms (racemers). All higher organisms use only the L-racemers. In fact, even bacteria use only L racemers for ordinary proteins. But they also use certain D-racemers in peptidoglycan. Does this suggest that the bacterial cell wall is older than the standard machinery of protein synthesis and harks back to a time when life wasn't so picky about which racemers it used? It could mean this. Certainly some scientists have thought so. But, since there are no similar structures in the Archaea or the Eukaryota, the more likely explanation is that this is a specialized feature of Eubacteria.
Perhaps, instead, these weird amino acids evolved as an ancient defense against attack by protein-digesting enzymes (proteases) which are efficient only in cutting ordinary peptide bonds. Notice the sequence of amino acids here: L-D-L-D. The region around the peptide bond [5] between amino acids will be neither left-handed nor right-handed, but a mixture likely to "confuse" proteases which are adapted to digest ordinary proteins. This kind of attack is most like to occur when a bacterium has been engulfed by a eukaryotic cell. So, the moral of this story may be that Eubacteria are not primitive forms at all, but specialized organisms which have been co-evolving with the eukaryotes for a very long time.
Bacterial cell walls contain a number of other typical components. However all of these are somewhat variable between different groups of bacteria; and we will defer discussion of these components for now. Briefly, the usual cell wall materials include teichoic acids, polysaccharides of various types, proteins and various derived lipids (fats). Teichoic acids may be of particular evolutionary interest, because they have elements of all of the major types of biomolecules. However, teichoic acids are found only in certain bacteria, and are not encountered outside the Eubacteria.
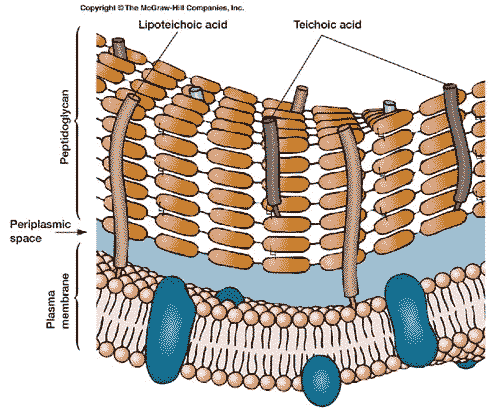 Moving inward from the cell wall, we encounter the plasma membrane. The bacterial cell membrane is, like virtually every other cell membrane, internal or external, based on a phospholipid bilayer. This basic structure of the membrane and wall together is shown in the figure on the left. To see why it forms a bilayer, we need only refer to the structure of a phospholipid. A phospholipid looks something like this:
Moving inward from the cell wall, we encounter the plasma membrane. The bacterial cell membrane is, like virtually every other cell membrane, internal or external, based on a phospholipid bilayer. This basic structure of the membrane and wall together is shown in the figure on the left. To see why it forms a bilayer, we need only refer to the structure of a phospholipid. A phospholipid looks something like this:
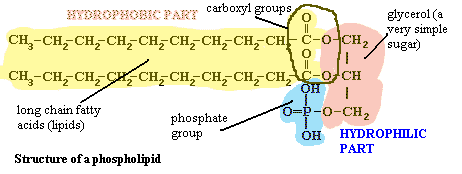
The "head" of the molecule contains a phosphate group, glycerol, and the carboxyl groups from two long-chain fatty acids. The details need not concern us, but it is important to recognize that these are all polar groups, groups that bind water tightly. That is, the head is hydrophilic. The long chain of carbons on the two "tails" of the molecule has no polar groups and so cannot bind water. These chains are hydrophobic. We might say that they are like oil in water. However, they are not simply like oil in water. That long featureless run of carbons is chemically identical to oil. With this kind of dual nature, the hydrophilic part will face the water and the hydrophobic part will exclude water. The most natural way to accomplish both tendencies is to form a bilayer, as shown in the image. See the footnote for a bit more detail [6].
The bacterial plasma membrane is a bit simpler than the plasma membrane of most eukaryotes. Eukaryotic cells have, in addition to phospholipids, cholesterol and other big, flat, mostly non-polar molecules which tend to stabilize the membrane and make it stiffer. Bacteria don't have cholesterol. Eukaryotes also frequently have a good many elaborate lipoproteins proteins with fats attached) and glycoproteins (proteins with sugars attached). These derivatized proteins do many of the same jobs which are performed by the cell wall and capsule in bacteria. Eukaryotes also have an extensive internal membrane system including the Golgi apparatus, endoplasmic reticula, vesicles, mitochondria, and a nuclear membrane. The Eubacteria have none of these. Some bacteria have small folds in the plasma membrane, in which some specialized functions may occur—notably the ATP-driven active transport of ions discussed above, as well as photosynthesis in the blue-green algae. However, there are no cytoplasmic membranes.
The cytoplasm itself is thus a rather uniform solution without a lot of structure. It does contain cytoplasmic inclusions of various kinds for storage of various critical metabolites. These include metachromatic granules of phosphate, glycogen (a polymer of glucose), grains of starch and salts, and poly(3-hydroxyalkanoate), the bacterial equivalent of fat.
In addition, the cytoplasm may contain plasmids. These are small, circular pieces of DNA derived from bacterial viruses (bacteriophages), other bacteria, or perhaps even other organisms. The genes carried on the plasmid may simply be dead weight, which continues to reproduce with the bacterium only because it happens to contain a working site for DNA polymerase. Thus, when this enzyme is active in the cell preparing the bacterial DNA for cell division, it tends to make a copy of the plasmid as well, even though the plasmid serves no biological function. On the other hand, plasmids can be of great practical importance, to humans as well as the bacterial host. Antibiotic-resistant disease bacteria often carry the extra genes which confer resistance on plasmids acquired from some completely different species. Plasmids may also contain other virulence factors, genes which code for proteins which can turn a harmless symbiotic species into a lethal disease vector.
As a starting point for comparison, let's briefly discuss DNA and what it does in metazoans—you, for example. We won't worry about the structure of DNA for now. Its enough to know that DNA is a polymer made of billions of nucleotides. Each nucleotide contains one of 4 bases. The sequence of bases is precisely the same in each cell of your body (with some exceptions for special cells), even though you have about 6 billion DNA nucleotides in each cell. Mitochondria and chloroplasts [7]have a bit of their own DNA. All of the rest is contained in a membrane-bound nucleus. This nuclear DNA is encased in chromatin,i.e., a regular coat of basic structural proteins (histones). The sequence of bases on the DNA is critical because it specifies a code for making proteins, as we'll discuss in a minute. However, eukaryotic DNA also contains very large stretches of DNA which serve other purposes: binding sites for regulatory proteins, recognition sequences for ligases, and many other functions, some unknown.
In the typical eukaryotic cell, RNA polymerases use DNA as a template to make long strands of RNA—a very similar polymer which has almost the same nucleotide/base structure. The process of making RNA from DNA is called transcription. The transcribed RNAs are spliced, recombined and "edited" in various steps in the nucleus, before being transported to the cytoplasm as mature messenger RNA (mRNA). In the cytoplasm, typically on the surface of an internal membrane system (the endoplasmic reticulum), the mRNA associates with ribosomes. Ribosomes are complex structures of protein and two molecules of ribosomal RNA, or rRNA. Ribosomes, and the rRNA molecules in them, are extremely conservative. The ribosomes of all eukaryotes are very similar. Because rRNA changes very slowly, rRNA is a good tool for looking at evolutionary relationships over huge lengths of time.
The ribosomes attach to the mRNA and use the sequences of RNA bases to make proteins according to the "genetic code." Proteins are polymers of amino acids. There are 20 amino acids commonly found in all organisms, and the sequence of amino acids determines what the protein is. Each run of three nucleotides on mRNA (each codon) specifies a particular amino acid. Since there are 4x4x4 = 64 possible codons, the code is redundant. That is, more than one codon sequence may specify a particular amino acid. This code is almost unbelievably conservative. Every living thing uses essentially the same code, with only a few, minor variations. See Table of Standard Genetic Code. This process of assembling proteins from the genetic code in mRNA is called translation. Translation is performed by the ribosomes using another type of RNA, transfer RNA or tRNA. There are 61 different tRNAs. Three codons are termination codons which signal the end of a coding sequence. Naturally, these have no tRNA. Each tRNA molecule has an anticodon at one end that binds to a specific mRNA codon. At the other end of the tRNA is the corresponding amino acid. The ribosomes move along the mRNA molecule, matching up the codons with tRNA anticodons, popping off the amino acid from the tRNA's far end, and adding the amino acid to the growing protein chain.
The bacterial chromosome is a single, circular molecule of DNA, vastly larger than any naturally occurring plasmid, but much smaller than almost all eukaryotic chromosomes. Bacterial DNA contains relatively few regulatory sites, by comparison with eukaryotic DNA, and has no extensive non-coding regions. Bacteria have no chromatin and, as mentioned above, there is no nucleus. The DNA is simply suspended in the cytoplasm. The Eubacteria use the same genetic code as metazoans. However, the bacterial ribosome, and ribosomal RNA, is considerably different. This may reflect a sharp functional difference. Bacterial DNA is not "edited" and transported. The ribosomes attach to the RNA even as it is being synthesized, so that transcription and translation are simultaneous and closely coupled.
This more or less concludes our whirlwind tour of cell biology and the bacterial cell. It is conventional to have lots of pictures and diagrams of transcription and translation. We have deliberately chosen to depart from this tradition in the interests of getting these painful necessities over quickly and without visual distraction. For good or evil, we will have plenty of opportunities for graphics later.
ATW030228
[1] Phytoplankton is an ecological term referring to any photosynthetic microorganism normally found in water, including forms (like Gomphosphaeria) that are often found attached to some substrate. The Cyanobacteria are the large group of very basal photosynthetic Eubacteria traditionally called "blue-green algae" because they use phycocyanin (a bluish pigment), as well as chlorophyll a (a green pigment), for photosynthesis of carbohydrates.
[2] Try it. Assume each cell is a cylinder 4 µ long, with a radius of 1 µ (1 µ = 10-6 m). Further assume a generation time of 1 hour and that the radius of the earth is 6.35 X 106 m. You can assume what you like about armpit height. It doesn't make much difference to the result.
[3] DO NOT get the idea that the bacterial cell has a net electrical charge. It doesn't. For every sodium (Na+) the cell pumps out, it must let some other positive charge in, typically potassium K+). This "battery" works by entropy, not enthalpy. I'll try to explain in a few sentences as follows. The pore cells are symmetrical. They grab onto any sodium—inside or outside the cell—change shape, then release the sodium and return to their original shape. There is no net gain or loss of potential chemical or electrical energy (or enthalpy). However, if there is much more sodium outside than inside, the net effect is that the transport works only one way, and can therefore perform useful work. How can this be? We seem to be getting work energy from nowhere!
In thermodynamics, at constant temperature, ΔG = ΔH - TΔS. ΔG is the change in free energy, energy that performs work. Doing work means that ΔG decreases. ΔH is the change in enthalpy, or (roughly) potential energy, including the energy stored in chemical bonds. If we break bonds, the ΔH decreases. T is the temperature measured from Absolute Zero, and ΔS is the change in entropy or the amount of disorder in the system. So, we can get useful work out of a system by reducing enthalpy (i.e. breaking bonds or discharging an electrical potential) or by increasing entropy (increasing the amount of disorder). By breaking down a concentration gradient, we increase entropy. The bacterial cell breaks down high-energy ATP molecules to create a sodium gradient, and so uses enthalpic potential energy to reduced entropy. Then it recovers its investment by letting the sodium back in, so increasing entropy, and converting that change in entropy to free energy used to turn the rotor.
[4] If you're familiar with plant cell walls, try to forget what you know. This is a completely different, and only distantly related, structure.
[5] The peptide bond is the -CONH- part that links two amino acids.
[6] Actually, the water excludes the long carbon chains, rather than the reverse. This turns out to be another entropy effect. In a bilayer, the non-polar lipid tails are do not interact with water and are free to be very disordered with respect to each other. The polar water molecules, like tiny temporary magnets, form temporary, shifting structures with their nearest neighbors. However, if a lipid molecule is forced into water solution, the water molecules still have no way to interact with the nonpolar tail. Instead, the water molecules form a sort of cage around the lipid, interacting with each other and creating a structure which has a good deal of long-range order. Worse, each water molecule near the tail has fewer neighbors, reducing the randomness of the interactions. Thus entropy strongly favors the bilayer. Even though it looks more ordered, the entropy of the entire system, including the water, is much higher when the long chains are only in contact with each other.
[7] Don't worry about what these are in detail either, if you aren't familiar with these organelles. Mitochondria are small, convoluted membranous structures in the cell which perform oxidative metabolism. That is, they use oxygen to break down sugars and related nutrients into water and carbon dioxide, using the energy released to create ATP for future use as en energy resource. Chloroplasts are layered membranous structures that perform photosynthesis. That is, they use energy from sunlight to build up sugars from carbon dioxide and water, releasing oxygen in the process. In essence, mitochondria and chloroplasts are opposites.
original page uploaded 24 March 2002
rewritten ATW030228
Last revised ATW030228
checked ATW060201, edited RFVS111214
this material may be freely used for all purposes
 This is the Domain of the Germs. The Eubacteria, in their hundreds of trillions, are the reasons you weren't allowed to pick up the candy you dropped on the floor or eat that egg salad that looked so good a week ago. They are behind every torture you have ever suffered at the hands of the dentist, and they are the root cause of childhood afflictions from antiseptics to acne. They have a lot to answer for. Then again, they're probably not too happy with us, either. Its hard to tell, since bacteria don't go in much for light conversation or email rants. They grow, or they don't, just as they have for the last three or four billion years, and without making much fuss about it. Bacterial psychology is thus a rather limited field. But such simplicity has its advantages. We are never tempted to paint human thoughts and emotions onto a 4 µ long pill-shaped blob of protoplasm. We can safely view the bacterium for what it is, a small biochemical machine, without having to steer the usual narrow passage between the twin perils of anthropomorphism and reductionism.
This is the Domain of the Germs. The Eubacteria, in their hundreds of trillions, are the reasons you weren't allowed to pick up the candy you dropped on the floor or eat that egg salad that looked so good a week ago. They are behind every torture you have ever suffered at the hands of the dentist, and they are the root cause of childhood afflictions from antiseptics to acne. They have a lot to answer for. Then again, they're probably not too happy with us, either. Its hard to tell, since bacteria don't go in much for light conversation or email rants. They grow, or they don't, just as they have for the last three or four billion years, and without making much fuss about it. Bacterial psychology is thus a rather limited field. But such simplicity has its advantages. We are never tempted to paint human thoughts and emotions onto a 4 µ long pill-shaped blob of protoplasm. We can safely view the bacterium for what it is, a small biochemical machine, without having to steer the usual narrow passage between the twin perils of anthropomorphism and reductionism.  This page will be devoted to considering the basic structural and functional units of the Eubacteria as biological machines. More specific matters, as well as considerations of ecology, phylogeny and evolution will be taken up in connection with more the specific groups to which these matters pertain. So, how are these machines put together? Some of the basic parts are shown in the figure on the left. Look at it carefully, because we have a good bit to cover, starting from outside.
This page will be devoted to considering the basic structural and functional units of the Eubacteria as biological machines. More specific matters, as well as considerations of ecology, phylogeny and evolution will be taken up in connection with more the specific groups to which these matters pertain. So, how are these machines put together? Some of the basic parts are shown in the figure on the left. Look at it carefully, because we have a good bit to cover, starting from outside. 
 Within the capsule is the relatively rigid
Within the capsule is the relatively rigid  Moving inward from the cell wall, we encounter the plasma membrane. The bacterial cell membrane is, like virtually every other cell membrane, internal or external, based on a phospholipid bilayer. This basic structure of the membrane and wall together is shown in the figure on the left. To see why it forms a bilayer, we need only refer to the structure of a phospholipid. A phospholipid looks something like this:
Moving inward from the cell wall, we encounter the plasma membrane. The bacterial cell membrane is, like virtually every other cell membrane, internal or external, based on a phospholipid bilayer. This basic structure of the membrane and wall together is shown in the figure on the left. To see why it forms a bilayer, we need only refer to the structure of a phospholipid. A phospholipid looks something like this: