
| Palaeos |  |
Glossary |
| Life | Glossary |
| Page Back: Evolutionary phylogeny | Page Up: Glossary | Unit home | Glossary (you are here) |
Page Next: References |
| Unit Back: Evolution | Unit Up: Life | Unit Down: (none) | References | Unit Next: Ecology |
16S ribosomal RNA or 16S rRNA) is a component of the 30S subunit of prokaryotic ribosomes. It is 1,542 nucleotides in length. Multiple sequences of 16S rRNA can exist within a single bacterium. The 16SrRNA gene is used for phylogenetic studies as it is highly conserved between different species of bacteria and archaea. Carl Woese pioneered this use of 16S rRNA. In addition to these, mitochondrial and chloroplastic rRNA are also amplified. (Wikipedia)
Actual common ancestor: term coined here to refer to the actual species that represents the common ancestor from which later species and groups evolved. Evolutionary Systematics is based on identifying and determining the traits of an ancestral species or, more usually, supra-specific taxa. Cladistics rejects the possibity of knowing the actual common ancestor, and instead posits a hypothetical common ancestor. (MAK120318)
Actual phylogeny: term coined here to refer to the actual evolutionary history of a group through deep time. Evolutionary Systematics, which is highly phylo-optimistic, seeks to reconstrtuct the actual evolutionary history of a group, including the actual common ancestor (either an ancestral species or, more usually, supra-specific taxa), and, with the help of the fossil record, tracing the evolution from ancestor to descendants and from there to further descendants . Cladistics, which tends to phylopessimism, denies it is possible to know the actual common ancestor, and instead posits a hypothetical common ancestor. (MAK120318)
Adams consensus: in cladistic analysis, a type of consensus method that uses the idea that a tree should be thought of as a "set of leaf subset nestings" rather than as a "set of clusters." A group nests within a larger group if the most recent common ancestor of the smaller group is a descendant of the most recent common ancestor of the larger group (from the PAUPDISPLAY Manual). This preserves all nested clades common to a set of source trees (Bininda-Emonds, 2004 - glossary) Adams consensus trees are designed to find the maximum number of components for a given set of cladograms by placing conflicting taxa at the most resolved node common to all the trees. (Forey et al 1992 pp.79-80). Only can be used for rooted trees. Usually preserves more structure than the strict methods, but may show clades in the consensus tree that do not occur in any of the trees in the set, which makes interpretation rather difficult. (from the PAUPDISPLAY Manual)
Advanced: see derived.
Algorithm: In mathematics and computer science, an effective method (a procedure that reduces the solution of some class of problems to a series of rote steps that give a specific and correct answer) expressed as a finite list of well-defined instructions for calculating a function. Algorithms are used for calculation, data processing, and automated reasoning. (Wikipedia)
Ancestor: in this context, an organism, or more correctly a population, lineage, or species, that through evolution gives rise to one or more descendants that generally belong to a distinct taxon or species to itself. The identification of ancestors and descendants is a central aspect of evolutionary systematics. In contrast, cladistics denies it is ever possible to know an ancestor (unless one can actually observe evolution in a laboratory). "No matter how well we understand our group, its taxonomy, paleontology and anatomy, we can never know if one taxon is ancestral to another" (Paraphyly Watch blog - Transitional Fossils, Microbes & Patrocladistics). See also Ancestral group, common ancestor. (MAK)
Ancestral group: I decided to adapt this phrase to refer to any supra-specific taxon or evolutionary grade which gives rise to another group. Examples include pelycosaurs, thecodonts, and condylarths. Ancestral groups are central to evolutionary systematics and often included in spindle diagrams. cladistics denies the validity of ancestral groups (see paraphyly). (MAK)
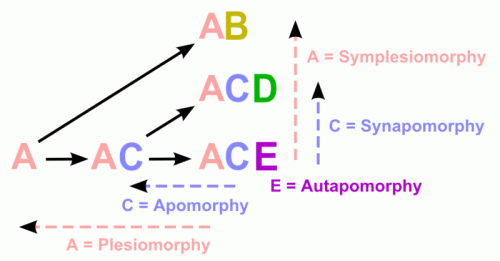
Apomorphy: In cladistics, an apomorphy is a unique derived or specialized character trait found in a particular taxon, which is also possessed by a common ancestor. character.
"A trait which characterizes an ancestral species and its descendants. This is an evolutionary novelty for the group. These are evidence for the existence of a group. Put another way, attributes shared in common are taken to indicate a shared evolutionary history.
A novel evolutionary trait that is unique to a particular species and all its descendants and which can be used as a defining character for a species or group in phylogenetic terms. Hence, the possession of feathers is unique to birds and defines all members of the class Aves. An apomorphy that is restricted to a single species is termed an autapomorphy. It alone cannot provide any information about the phylogenetic relations of that species, although it can indicate the degree of divergence of a species from its nearest relatives. An example is speech, which is found solely in humans (Homo sapiens) and not in other primates. An apomorphy that is shared by two or more species or groups is termed a synapomorphy. Such traits define the strictly monophyletic groups, or clades, which are the basis of cladistic classification systems (see cladistics). Compare plesiomorphy."
A Dictionary of Biology, Oxford University Press, © Market House Books Ltd 2000
In phylogenetic nomenclature, an apomorphy-based clade is a clade the members of which are defined through their possession of that particular trait. Contrast with autapomorphy, an apomorphy found only in a single taxon and of no phylogenetic (cladistic) value.
Apomorphy-based taxon (or clade): a group comprising all species descended from a common ancestor characterised by specific apomorphies. Apomorphy-based taxa are rarely used in cladistics because of the difficulty of determining when a particular trait appeared and whether its presence can be reliably determined. In contrast, all Linnaean and Evolutionary systematic taxa are apomorphy-based (either paraphyletic or monophyletic). Contrast with node-based and stem-based MAK120318
Artefact: not an ancient extraterrestrial or interdimensional device of great power, but, in the more mundane phylogenetic systematics and phylogenomic context, a false signal resulting in a distorted phylogenetic view of the group being studied. Examples include Long Branch Attraction and Heterotachy. See also Garbage in, garbage out.
ASCII Phylogenetic Tree: As here defined, an ASCII phylogeny, or more correctly an ASCII Phylogenetic Tree, is a dendrogram or tree diagram which uses ASCII-text format to draw supertrees. ASCII Phylogenetic Trees might also informally be referred to as ASCII Cladograms, but that is inaccurate because cladograms are, pragmatically speaking, not actually phylogenies but branching diagrams depicting patterns of shared similarities (O'Keefe & Sander 1999). The ASCII Phylogenetic Tree format was created by T. Mike Keesey, who used them to show dinosaur phylogenetic relationships in the old Dinosauricon. Mikko Haaramo adopted this format, but refined it with the introduction of the grave ( ` ) for the corners, for his own phylogenetic archive. This useful format was then adopted on the Dinosaur Mailing List and by paleo enthusiast webmasters like Jack Conrad (The Vertebrate Phylogeny Pages), Justin S. Tweet (Thescelosaurus!), Øyvind M. Padron (The Dinosauria), and Toby White and myself (Kheper palaeo and Vertebrate Notes, and finally Palaeos.com, here). The format has now become pretty standard in any paleo geek text-based phylogenetic diagram. (MAK)
Autapomorphy: a character traitunique to a particular unique to a particular taxon; Because autapomorphies do not provide information about the organism's phylogenetic relationships to other taxa, they are of no use in cladistics. However they still provide useful non-phylogenetic information about the species in question. Compare with apomorphy, plesiomorphy, homology, homoplasy.
Basal: Preferred cladistic substitute for "primitive", as it is felt the latter may carry false connotations of inferiority or a lack of complexity. In cladograms, basal taxa are those terminal taxa that first diverged from the root. The term basal is only be correctly applied to clades (species or higher groups) of organisms, not to individual traits possessed by the organisms. There is however a tendency for terms like basal and stem to appear as rather vague alternatives to "primitive" or "ancestral" in cladistic paleontological literature and especially popularised accounts and comments thereof (MAK120318, Wikipedia)
Basal node: the node or base of the cladogram, representing the hypothetical common ancestor of the entire clade (however if the common ancestor or something like it is known, then it is shown as a terminal taxon, see basal taxon. See graphic. (MAK)
Basal taxon: general term in phylogenetic systematics for any terminal taxa that lie at the base of a cladogram, i.e. they are connected by, or else close to, the basal node, and their sister group is the sub-clade that constitutes the rest of the cladogram. Equivalent to primitive or ancestral (these terms not being used in cladistics). Included under or partially equivalent to stem group. (MAK)
Bayesian inference: a method of statistical inference, used for example in cladistics algorithms, in which some kinds of evidence or observations are used to calculate the probability that a hypothesis may be true, or else to update its previously calculated probability.
Binomial nomenclature: Linnaean universal standard of biological scientific notation, according to which every species is given a distinct two-part name. The first part, think of it as like the surname, is the genus, which is capitalised, the second part the species, written completely in lower case, is like the given name. Both names are by convention written in italics (or if that is not possible, underlined, or if even that is not possible say with ASCII text, then there is an underscore character before and after the name, _like this_). So in the case of Tyrannosaurus rex, Tyrannosaurus is the genus (capital "T"), and rex (small "r") the species. Finally, the name of the discoverer of the species is added (if the species has since been given a new genus the discoverer's name is placed in brackets) along with the year of publication of the scientific paper describing that particular species. Hence (etc). This usage is not mandatory in popular and semi-technical books, but is when describing or listing species in a technical journal or a Museum. The species name can also be abbreviated by only using the first letter of the genus and a period, after which comes the species name. the species name on its own can be written as T. rex (but never "T-rex", it's not a car!). Any student of natural history will be familiar with this approach. I have noticed however a tendency among paleontologists to give every new discovery a new genus as well as a new species, leading to an over-excess of monotypic genera (each genus only having one species). This was and is exacerbated by the cladistic revolution, where even species previously placed in a genus are moved to their own genus, especially if precise phylogenetic relationship is uncertain (which it almost always is in these cases) only adding to the multiplication of names (Paleo artist and author Greg Paul at one time (Predatory Dinosaurs of the World, 1988) went in the opposite direction, lumping species from even fairly distant genera together; e.g. most dromaeosaurids became Velociraptor, although his more recent work The Princeton Field Guide to Dinosaurs (2010) provides a nice middle of the road balance). There is also a move among proponents of phylogenetic nomenclature and the Phylocode to abandon the binomial altogether, and emphasise only the phylogenetic relationships (which wouldn't necessarily be evident from the name alone). This would actually only be a small step when considering vertebrate paleontology alone in view of the above mentioned tendency (but not for example Pleistocene mammals which have a very good fossil record!), but would be a nightmare if cataloging or referencing all of the other millions of named and described species. However it is probably unlikely that the Phylocode will become a majority position any time soon. (MAK)
Bootstrap - a sampling method used in cladistics. Boostrapping measures how consistently the data support a given tree topology. It is not determine how accurate a cladogram is; it only gives information about its stability, and helps assess whether the sequence data is adequate to validate the topology (branching order) (Holmes 2003)
Bremer support: The Bremer support for a clade is the number of extra steps you need to construct a tree (consistent with the characters) where that clade is no longer present. There are reasons to prefer this index rather than the bootstrap value. (Øyvind Hammer - PAST - Paleontological Statistics Software) (see also decay index)
Bubble diagram: Informal neologism used by yours truly for a spindle diagram with rounded rather than angular contours. Also called a romerogram. (MAK)
Character, Character State: any recognizable trait, feature, or property of an organism, used to reconstruct phylogenies. Characters may be morphological, behavioral, physiological, or molecular. In cladistics, the character is thought to be derived and vary from a corresponding feature in a common ancestor of the organisms being studied. (modified from PBS evolution Glossary and UCMP Understanding Evolution Glossary, also definitions from Sereno, 2007)
Character State: the mutually exclusive conditions of a character. (one of the possible alternative conditions of the character). For example, "present" and "absent" are two states of the character "hair" in mammals. Together, characters and character states compose what are termed character statements. (modified from PBS evolution Glossary and UCMP Understanding Evolution Glossary, also definitions from Sereno, 2007)
Character state change: change of the form or state of a character in the course of evolution.
Character optimization, character mapping: interpreting characters on a phylogenetic tree in order to reconstruct ancestral character states.
Chronogram: phylogenetic tree that explicitly represents evolutionary time through its branch lengths. (Wikipedia)
Chronospecies: One or more species which continually changes from an ancestral form along an evolutionary scale. This sequence of alterations eventually produces a population which is physically, morphologically, and/or genetically distinct from the original ancestors. Throughout this change, there is only one species in the lineage at any point in time, as opposed to cases where divergent evolution produces contemporary species with a common ancestor. Relies on an extensive fossil record, since morphological changes accumulate over time and two very different organisms could be connected by a series of intermediaries. The related term paleospecies indicates an extinct species only identified with fossil material. To avoid unnecessary multiplication of terminology (and paleontology-neontological distinctions) these terms are here synonymised. For example, changes in the Permian lepospondyl amphibian Diplocaulus over time may imply a chronospecies (= paleospecies). (MAK, Wikipedia)
Clade: term coined by Julian Huxley, in terms of evolutionary branching and ancestry, to refer to the set of all organisms descended from a particular ancestor. In cladistics, a clade is a monophyletic group of organisms that includes all the descendants of a common ancestor as well as that ancestor itself. For example, birds, dinosaurs, pterosaurs (flying reptiles), crocodiles and their extinct relatives all form the clade Archosauria. In phenotype-based Linnaean and evolutionary systematics, clades are not always suitable as units of classification, as the crown portion of a clade may be very different from its base (compare a pelycosaur reptile to a eutherian mamal for example). The Phylocode attempts to formalise phylogenetic systematic taxonomy based on the use of clades. Contrast grade. (MAK)
Cladistic species concept: based on cladism, this is a definition of a species as a lineage of populations between two phylogenetic branch points (or speciation events). But because most speciation is through budding rather than cladogenesis, this definition is either problematic, or needs to be modified. Compare with biological species concept, ecological species concept, phenetic species concept, and recognition species concept. (Fossil Mall glossary, MAK)
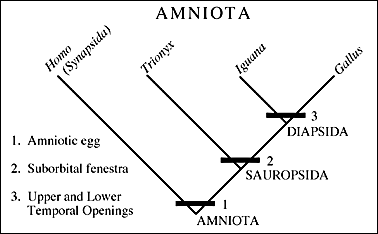 Cladogram by Paul Olsen (original url), showing four species (human, turtle, lizard and bird) and three clades, each defined buy its own synapomorphies (shared unique characteristics). Most cladogram matrixes involve many hundreds of such characteristics |
Cladistics: Rigorous methodology first developed by Wili Hennig, to evaluate and reconstruct phylogenetic hypotheses. The results of cladistic analyses are often represented in the form of a branching diagram, called a cladogram. Note that cladistics is not the same as phylogeny, and cladograms are not phylogenetic diagrams of ancestor-descendant relationships! As with Linnaean classification, cladistics provides a nested hierarchy where an organism is assigned a series of names that more and more specifically locate and define it within the hierarchy. However, unlike Linnaean classification, phylogenetic classification only allows monophyletic clades, excludes both paraphyletic and polyphyletic groups. It also does not assign ranks (e.g. class, phylum) to the hierarchical levels.
The early 1980s saw conflict between the two schools of Pattern Cladism and Hennigian systematics, although this has since been resolved, and cladistics today is generally based equally on both. In the 1980s and 90s cladistics became the dominant paradigm in biological systematics, supplanting the previous Linnaean-based evolutionary taxonomy in all fields except botany. Together with molecular phylogeny it forms the current Phylogenetic paradigm.
Despite being often referred to as "phylogeny", cladistics does not seek to describe the actual course of phylogeny in deep time a la evolutionary systematics, but only selecting the most viable hypothesis regarding phylogenetic relationships, given the available data (empirical method). In other words, cladistics in general tends towards phylopessimism rather than phylo-optimism
It is held by cladists that taxa (if recognized) must always correspond to clades, united by apomorphies (derived traits) which are discovered by a cladistic analysis. To this end, cladistics collects character data only from the taxa being studied, and do not consider the inferred characters of ancestors. With the advent of powerful computation, cladistics uses statistical procedures such as Bayesian analysis and Maximum Likelihood. Such computer-based cladistics has been especially popular in paleontology. (MAK) ATW - Introduction to Cladistics ; Links: What is cladistics? How reliable is it? - Mike Taylor; Cladistics - Wikipedia; 29+ Evidences for Macroevolution: Phylogenetics - Douglas Theobald @ Talk Origins. More
Cladogenesis: The division of an ancestral parental lineage into two or more daughter lineages or species. At one time, cladogenesis was recognised, along with anagenesis, as one of the two types of gradual evolution. Because the highly formalised trees that cladistics use on do not show anagenesis, a misplaced literalism led to cladogenesis, in the sense of the division of a common ancestor into two daughter species, being accepted as the standard form of speciation. However, other evolutionary processes, especially budding and merging, involve asymmetrical divergence and therefore paraphyly. (MAK)
Cladogram: A dichotomous phylogenetic tree that branches repeatedly, suggesting a classification of organisms based on the sequence in which evolutionary branches arise; a nested diagram of synapomorphies indicating relations between groups; each point of branching represents divergence from a common ancestor. In the 1980s, cladograms were originally based on immediately apparent synapomorphies. From the 1990s onwards, computational phylogenetics are now very commonly used in the generation of cladograms. Characters pertaining to each taxon are run through computer algorithms to determine phlogenetic relationships. Although traditionally cladograms were generated largely on the basis of morphological characters alone, nowadays DNA and RNA sequencing data have been used as well. All have different intrinsic sources of error. For example, character convergence (homoplasy) is much more common in morphological data than in molecular sequence data, but character reversions that are unrecognizable as such are more common in the latter (see long branch attraction). Morphological homoplasies can usually be recognized as such if character states are defined with enough attention to detail. The researcher must decide which character states were present before the last common ancestor of the species group (plesiomorphies) and which were present in the last common ancestor (synapomorphies) and does so by comparison to one or more outgroups. The choice of an outgroup is a crucial step in cladistic analysis because different outgroups can produce trees with profoundly different topologies. Note that only synapomorphies are of use in characterizing clades (Wikipedia). Contrary to popular belief, cladogram nodes do not represent actual ancestral taxa. Were an actual ancestor to be included it would ideally appear (if the cladogram is correct in this regard) as the sister taxon of the sub-clade that includes all its descendants. (MAK)
Class: In the Linnaean classification the taxonomic rank between phylum and order, used to define major sub-group within a phylum. Classes are used in the taxonomic series of evolutionary systematics but are not used in cladistic analysis., Classes are often paraphyletic. This is shown by the spindle diagram showing the evolution of the vertebrates, where only five out of nine classes are holophyletic clades. However this is due to the fact that cladistics uses the species as its basic reference point, whereas evolutionary systematics tends to use families, orders, classes, and phyla. (MAK) More
Coalescent Theory: A method for comparison of gene sequences in populations to find the most likely common ancestor sequence. (W. R. Elsberry - talk.origins)
Common ancestor: The ancestral species that gave rise to two or more descendant lineages, and thus represents the ancestor they have in common. The idea of a common ancestor is central to evolutionary thinking from Darwin onwards. In the Modern Synthesis' Evolutionary Systematics the common ancestor is usually shown as the most suitable fossil form at the base of a lineage, where it may or (more likely given the small number of species known from those which actually lived in past ages) or may not be an actual ancestor, more often it is a sort of grand-uncle rather than grandfather). Nevertheless, some idea of a general common ancestor can be had. In an attempt to establish greater rigour and precision, Cladistic phylogeny defines the most recent common ancestor as the originator of a clade; in other words the first species or organism to possess the unique attributes of that clade. Contrary to popular opinion, cladograms do not actually show the actual common ancestor; in this context, see basal taxon, hypothetical common ancestor. See also non-missing link. (MAK)
Computational phylogenetics: the application of computational algorithms, methods and programs to phylogenetic analyses. The goal is to assemble a phylogenetic tree representing a hypothesis about the evolutionary ancestry of a set of genes, species, or other taxa. A central element in modern cladistics. (MAK, Wikipedia)
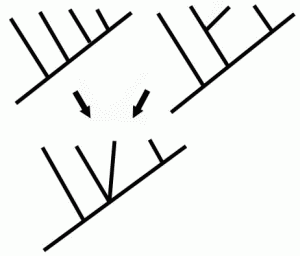
Consensus: in cladistics, a consensus tree is the agreement between two or more trees (see diagram at right). Obviously there are many different possible solutions, as well as different methodologies. Some consensus methods include strict, majority rule, semi-strict, Nelson, and Adams consensus.
Graphic: A Consensus Cladogram, from How We Look at the Relationships of Taxa
Consistency index (CI): In cladistics, the measure of the parsimony fit of a character to a tree, or of the average fit of all characters to a tree. Varies from 1.0 (perfect fit) to a value asymptotically approaching zero (poorest fit). It is inflated by autapomorphies which can only take the value 1.0; thus a totally uninformative data set (consisting only of autapomorphies) could return a CI equal to 1.0. Compare retention index. (Michael D. Crisp - Introductory glossary of cladistic terms). The per-character consistency index (ci) is defined as m/s, where m is the minimum possible number of character changes (steps) on any tree, and s is the actual number of steps on the current tree. This index hence varies from one (no homoplasy) and down towards zero (a lot of homoplasy). The ensemble consistency index CI is a similar index summed over all characters. (Øyvind Hammer - PAST - Paleontological Statistics Software)
Crown group: in cladistics, a group consisting of living representatives, their ancestors back to the most recent common ancestor of that group, and all of that ancestor's descendants. The name was given by Willi Hennig as a way of classifying living organisms relative to extinct ones. Though formulated in the 1970s, it was not commonly used until its reintroduction in the 2000s. The usual definition of a crown group is the smallest monophyletic group, or "clade", to contain the last common ancestor of all extant members, and all of that ancestor's descendants. Extinct side branches on the family tree will still be part of a crown group. For example, if we consider the crown-birds (i.e all extant birds and the rest of the family tree down to their last common ancestor), extinct side branches like the dodo or great auk are still descended from the last common ancestor of all living birds, so falls within the bird crown group. (MAK, Modified from Wikipedia)
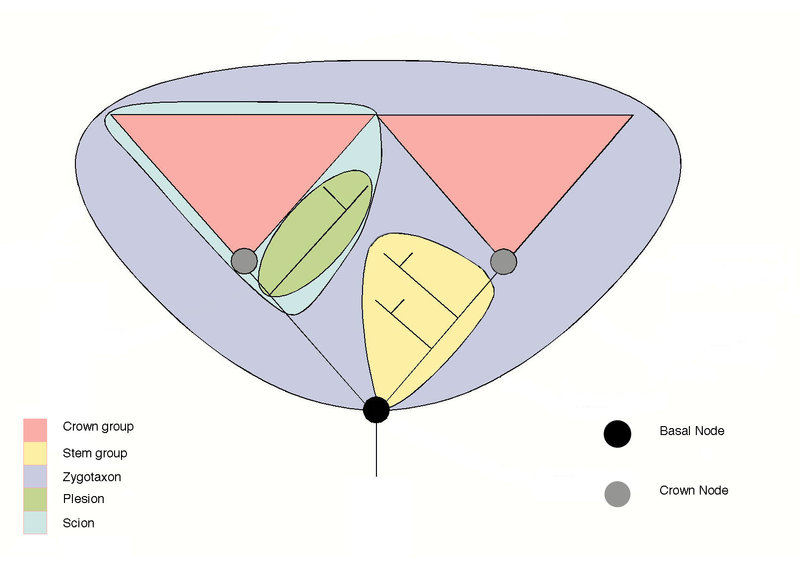 From Wikipedia. The stem and crown group concept. The two pink groups represent a pair of crown groups, the last common node of which is the basal node. Terminology is from Craske, A. J. and Jefferies, R. P. S. (1989) A new mitrate from the late Ordovician of Norway, and a new approach to subdividing a plesion. Palaeontology 32, 69–99 and Budd, G. E. (2001) Tardigrades as "stem-group" arthropods: the evidence from the Cambrian fauna. Zoologischer Anzeiger 240, 265-279. Diagram and text by Graham Budd. The diagram shown here is revised from the original to clarify that the stem group does not include the basal node (ancestor) of the crown group. text and revision by Peter Coxhead. For explanation of terminology see Wikipedia - Crown Group page. |
Daughter group: see Sister group.
Decay index: In cladistic analysis, the number of additional steps required to dissolve a given clade (Michael Allaby, 1999, Dictionary of Zoology). (see also Bremer support)
Dendrogram: There doesn't seem to be an agreed meaning of this term. Michael Crisp's cladistic glossary defines it as any branching diagram or tree, such as a cladogram. Mayr & Bock 2002(for the evolutionary systematics camp) contrast the "Hennigian cladogram" with the "Darwinian dendrogram". The Wikipedia page gives another definition again: "a tree diagram frequently used to illustrate the arrangement of the clusters produced by hierarchical clustering. Dendrograms are often used in computational biology to illustrate the clustering of genes or samples." As used on Palaeos, a dendrogram is any informal phylogenetic cladogram-like diagram, a sort of composite of published trees, or simply the author in question's random opinion. MAK111018 120318
Derived: same as apomorphy; a derived character / trait is inferred to be a modified version of a more primitive condition of that character and therefore inferred to have arisen later in the evolution of the clade.
Descendant: in this context, a population, lineage, or species, that arises through evolution from an ancestor (an earlier species or taxon). Where a number of descendants share the same ancestor (cladogenesis), the ancestor is called a common ancestor. (MAK)
Distance: phylogenetic or evolutionary divergence. Distances are usually expressed pair-wise among terminal taxa, and can be calculated based on a specified evolutionary model; the model specifies the probabilities of character-state changes through evolutionary time. Distances are popular for building phylogenetic trees from molecular sequence data Compare with maximum likelihood, parsimony. (Michael D. Crisp - Introductory glossary of cladistic terms)
Doubly paraphyletic group: a group or taxon that is paraphyletic because two of its descendant lineages are not included. e.g. Class Reptilia is a doubly paraphyletic group because reptiles seperately evolved into birds and mammals. cf. singly and triply paraphyletic groups (MAK120318)
Electrophoresis: The method of distinguishing entities according to their motility in an electric field. In evolutionary biology and molecular sequencing, it has been mainly used to distinguish different forms of proteins. The electrophoretic motility of a molecule is influenced by its size and electric charge. (PBS evolution Glossary)

Elvis taxon: a taxon which has been misidentified as having re-emerged in the fossil record after a period of presumed extinction, but is not actually a descendant of the original taxon, instead having developed a similar morphology through convergent evolution. This implies the extinction of the original taxon is real, and the two taxa are polyphyletic. The term was coined by D. H. Erwin and M. L. Droser in a 1993 paper to distinguish descendant from non-descendant taxa: "Rather than continue the biblical tradition favored by Jablonski [for Lazarus taxa], we prefer a more topical approach and suggest that such taxa should be known as Elvis taxa, in recognition of the many Elvis impersonators who have appeared since the death of The King." Lobothyris subgregaria, a brachiopod from the early Jurassic period, is one example of such a taxon. By contrast, a Lazarus taxon is one which actually is a descendant of the original taxon, and highlights missing fossil records, which may be filled later. A Zombie taxon is a taxon sample that was mobile in the time between its original death and its subsequent discovery in a site of younger classification, like, for example, a trilobite that gets eroded out of its Cambrian-aged limestone matrix, and reworked into Miocene-aged siltstone. (Wikipedia)
Evolutionary classification: see Evolutionary systematics
Evolutionary clock: see Molecular clock
Evolutionary phylogeny: informal name for a synthesis of evolutionary systematics, cladistics, molecular phylogeny, stratocladistics, and anyhing else worth throwing into the mix, based on the premise that no one methodology alone can give all the answers MAK120318 More
Evolutionary systematics, also called Evolutionary classification, a way to determine natural relationships of organisms by studying a group in detail and comparing degree of similarity. Tends to consider supra-specific taxa rather than single species. The origin of a major new trait or apomorphy (e.g., flowers in angiosperms, endothermy and lactation in mammals) results in the formation of a new "natural group" of the same Linnaean rank as the "natural" group from which it arose (in these examples gymnosperms and reptiles respectively). Often uses spindle diagrams that map taxonomic diversity (usually mapped on the horizontal axis) against geological time (mapped vertically, in keeping with the geologists' tendency to equate time with geological strata and hence verticality). Evolutionary systematics has its roots in the work of Haeckel, but reached its current form in the modern evolutionary synthesis of the early 1940s, especially the work of George Gaylord Simpson and Ernst Mayr. In this school of thought, classification reflects both phylogenetic relatedness as well as morphological disparity (overall similarity). (MAK, Wikipedia, Grant, 2003, IAB blog, quoting Ebach et al 2008, UCMP Virtual Paleobotanical Laboratory) More
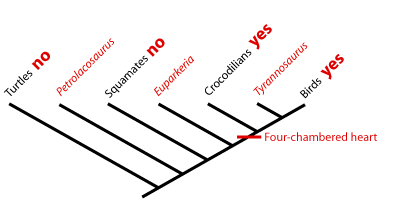
Extant Phylogenetic Bracket; Phylogenetic bracketing: In 1999, Larry Witmer described how unknown character states for fossil taxa are reconstructed with respect to extant taxa called the extant phylogenetic bracket (EPB). This is the bracket formed on either side of the taxon with the missing information by extant taxa in which the character state is known. Using it, we can make three types of inference, listed in order of decreasing confidence. Consider the distribution of a soft-tissue character - the four-chambered heart - among three fossil reptiles:
- Type I Inference: Tyrannosaurus is bracketed by birds and crocodilians, both of which have the derived character. With no contrary positive evidence, the simplest assumption is that Tyrannosaurus had it also.
- Type II Inference: The basal archosauriform Euparkeria is bracketed by crocodilians and squamates. Crocs have the derived character, squamates don't. Thus, we are much less secure than above in inferring it in Euparkeria , but presence of some sort of hard tissue correlate of that trait might increase our confidence.
- Type III Inference: The basal diapsid Petrolacosaurus is bracketed by squamates and turtles, neither of which have the derived character. Our confidence in its presence in the extinct form is very low. We would need strong positive fossil evidence to argue for its presence.
Text and diagram by John Merck
Family: In the Linnaean classification the taxonomic rank between order and genus (or order and tribe, tribe being a mostly botanical rank between family and genus), used to define group of related organisms. Used in evolutionary systematics but not cladistics. (MAK) More
Five Kingdoms: evolutionary classification of life developed by Robert Whittaker and Lynn Margulis, according to which organisms are divided into five kingdoms: Monera, Protist, Plants, Fungi, and Animals. More
. "Gap codings". this is not a formal term but refers to the situation in cladistics, when a 'daughter' character is logically dependent upon the state of a 'parent', and cannot be coded when the parent is absent. For example, the position of the frontal appendage in an arthropod can only be coded in taxa that possess a frontal appendage in the first place. In morphological analyses, this assigns double weight a priori to absences in the 'parent' character (because the daughter is always contingent, that is, dependent on the parent character), and can artificially inflate support for particular clades, and hence affect overall tree topology. This situation is hard to avoid when selecting characters across a range of fossils, which include taxa with unusual or differing morphologies. In analyses of nucleotide data the situation is different, because gaps may be the result of shared deletions from an ancestral sequence and hence be informative. (Mounce&Wills2011, Liuetal2011)
"Garbage in, garbage out": self-explanatory phrase borrowed from computer programming. If the characters used in phylogenomics (and cladistic analysis in general) are unreliable, even the most accurate tree reconstruction method can fail. Therefore, methods focusing on the most reliable characters have been developed in order to reduce the impact of inconsistency. (Delsuc et al 2005)
Genealogy: Term derived from Greek γενεά, genea, "generation"; and λόγος, logos, "explanation". The study of families and the tracing of their lineages and history. Genealogists use oral traditions, historical records, genetic analysis, and other records to obtain information about a family and to demonstrate kinship and pedigrees of its members. The results are often displayed in charts or written as narratives. In evolutionary thought, such as cladistics, due to the alternate translation of γενεά as "race" the term can be used as a synonym for phylogeny. (from Wikipedia, Perseus Digital Library, revised RFVS111126)
Genetic Algorithms: Computational systems based upon an implementation of natural selection as an algorithm for classification or optimization. (W. R. Elsberry - talk.origins)
Genus: In the Linnaean classification the taxonomic rank between family or tribe and species, and used to define group of closely related organisms that differ in only very minor ways. In the Linnaean system of binomial nomenclature, the genus is written in italics, with a capital letter, in front of the species name, or on its own. e.g. with Tyrannosaurus rex, the name Tyrannosaurus is the genus, and T. rex (no hyphen!) is the species. Used in evolutionary systematics; in cladistic classification every genus is only allowed two species (because of excessive formalism regarding cladogenesis), and Linnaean genera are always oversplit and new names created, resulting in much taxonomic confusion (for example in paleontology the established dinosaur genus Iguanodon has been split into about a dozen different monospecific genera (link). See also the discussion at Sauropod Vertebra Picture of the Week. It may be that the Phylocode will discard binomial nomenclature altogether (although there is obvious resistance to this) . (MAK) More
Ghost lineage: in cladistics, a phylogenetic lineage that is inferred to exist, for example by matching a cladogram against geological time, but is not known from the fossil record.
When we know that two taxa are sister taxa (descendants of the same recent common ancestor), we in essence know that they originated at the same point in geologic time - the time of their last common ancestor and the speciation event that gave rise to them. Say we know one of these taxa from 100 million year old rocks, and the other from 90 million year old rocks. Even without seeing a fossil, we know that the second group must have representatives dating back at least to 100 million years, simply from its sister-taxon relationship with the other. A lineage like this, whose existence can be inferred from the cladogram, but which is not known from actual fossils is called a ghost lineage. The examination of ghost lineages should allow biostratigraphers to refine their models of the stratigraphic ages of organisms.
Links: UCMP, evowiki, Dave Hone's Archosaur Musings (MAK)
Grade, Evolutionary: a paraphyletic group showing similarities in morphology, ecology or life history; a horizontal taxon consisting of transitional forms between two other taxa. (MAK). In alpha taxonomy, a grade refers to a taxon united by a level of morphological and/or physiological complexity. The term was coined by British biologist Julian Huxley, to contrast with clade, a strictly phylogenetic unit. (Wikipedia)
Gradism; Gradistics: as used here, the opposite (or complementr) of cladistics; understanding phyloigeny in terms of evolutionarey tramnsformation and ancestor-descendent relationships. Includes the evolutionary systematics of .
Great chain of being: metaphysical premise, popular from the classical world until the early 19th century, that all beings constitute a single continuous series of forms in an unbroken gradation from God through countless intermediate spiritual and material stages to formless matter. Also used more profanely to justify feudalism, the Church, etc. With the development of naturalistic theories of evolution and phylogeny, representations of the great chain of being was replaced by phylogenetic trees and secular cosmology, although it remains popular outside science, where it is interpreted as "ascent". See also The March of Progress. (MAK)
Heterotachy. Variation in the evolutionary rate of a given position of a gene or protein through time. Can lead to phylogenetic reconstruction artefacts where unrelated taxa have converged in their proportions of invariable sites. Unlike other types of bias, heterotachy does not leave any evident traces in sequences, and therefore are particularly difficult to detect. (Delsuc et al 2005)
Holophyletic, Holophyly: Ashlock 1971 coined the term to resolve the ambiguity between the Haeckelian (evolutionary systematic) and Hennigian (phylogenetic systematic, cladistics) definitions of monophyly, and that usage is followed here. Refers specifically to the definition that a group contains the common ancestor, all organisms descended from the common ancestor, and no other organisms. The term has not gained widespread acceptance in the scientific community, probably because monophyletic is so widely used and has the same meaning. (MAK, Wikipedia)
Horizontal classification: as described by Simpson, , a taxon based on overall similarity between its members at a particular time. All members share a common ancestry and are therefore monophyletic at that time slice, however, only the members extant at that particular time are considered. An evolutionary grade. Evolutionary systematics includes the interplay of both horizontal and vertical classification, whereas cladistics only considers vertical. (MAK) More
Hypothetical common ancestor: it is necessary to distinguish between cladistics and evolutionary systematics, as the two tend to confused in a sort of mishmash in the popular imagination and on some Wikipedia diagrams. In contrast to the evolution trees (spindle diagrams and so on) that evolutionary taxonomists use, cladograms are not intended to portray actual phylogeny. i.e. a cladogram does not have a time axis, and it does not portray ancestors, but only branching patterns, that is, sister relationships between terminal taxa and other nodes. This means that the internal nodes that lie at the base of each nested clade do not represent an actual species which can be described in terms of traits and characters, but rather a hypothetical and abstract representation of the common ancestor of that particular clade. (MAK)
International Code of Zoological Nomenclature: widely accepted convention in zoology that rules the formal scientific naming of organisms treated as animals. The rules principally regulate:
The rules and recommendations have one fundamental aim: to provide the maximum universality and continuity in the scientific naming of animals. The code is published by the International Commission on Zoological Nomenclature (ICZN), an organization dedicated to "achieving stability and sense in the scientific naming of animals". The rules in the Code determine what names are valid for any taxon in the family group, genus group, and species group. It has additional (but more limited) provisions on names in higher ranks. Several cladists have argued that the Linnaean based ICZN code needs to be replaced by a new cladistically-based system, the Phylocode. (Wikipedia)
Intuition: in this context, arriving at a scientific (or any creative) hypothesis through a leap of insight. For example, Einstein discovered Special Relativity by imagining what it would be like to ride on a photon. From another perspective, gut-feelings, hunches, creativity, and more. See also art. In systematics, advocates of Phenetics and Cladistics argue on pragmatic grounds that evolutionary systematics should be rejected because it is too "intuitive", and not sufficiently verifiable. However their use of quantitative empirical data without intuition meant they were not able to distinguish homology from homoplasy. Hence all science will always include some intuition and subjectivity. (MAK)
Junior synonym: giving a new name to a species, supra-specific taxon, or clade which already has a scientific name. As a standard, the first applied name is the one that is used in biological and paleontological systematics. Junior synonyms are redundant and hence usually rejected in scientific nomenclature; the exception being when the more recent name is so well known that to change it would cause confusion. For example, the first named fossil which can be attributed to Tyrannosaurus rex consists of two partial vertebrae found by Edward Drinker Cope in 1892 and named Manospondylus gigas. It was only later realised that they belong to the same animal. In this case, the newer name, Tyrannosaurus rex (named by Henry Fairfield Osborn in 1905) was retained, and the older one Manospondylus gigas, rejected. (MAK, Wikipedia)
Kingdom: In the Linnaean classification the highest taxonomic rank. Traditionally only included plants and animals; Whittaker-Margulis classification scheme adds three more kingdoms, and other researchers such as Thomas Cavalier-Smith have added additional kingdoms.
Last (or Latest) Common Ancestor (LCA) the most recent common ancestor of any two (or more) species, which is another way of saying it is the earliest member of a particular clade that includes those species but not more distantly related species. So there are still earlier ancestors, and they would also be common ancestors, but they would include other taxa as well as those being studied, and would stand at the base of a more inclusive clade. e.g. the most recent common ancestor of a dog, a cow, a human and a chimpanzee (Boreoeutheria) is also the common ancestor of a human and a chimp (Hominidae), but it isn't the most recent one (it lived much earlier and evolved into far more groups of animals). Therefore, to limit study to the group or clade under consideration, only those members in that clade, and their most recent (not oldest) common ancestor is considered. (MAK120318)

Lazarus taxon: a taxon that disappears from one or more periods of the fossil record, only to appear again later. An example is Lazarussuchus, an Oligocene member of a clade of freshwater reptiles (Choristodera) thought to have gone extinct at the end of the Mesozoic. As Lazarussuchus is thought to be outside the clade including other choristoderans, it may indicate a ghost lineage going back to the Late Triassic, a span of over 170 million years. There are also examples of "Burgess Shale type fauna", best known from the Early and Middle Cambrian periods, but which, since 2006, have been found in rocks from the Ordovician, Silurian and Early Devonian periods, in other words up to 100 million years after the Burgess Shale (Kühl et al 2009; Siveter et al 07). The term "Lazarus taxon" refers to the account in the Gospel of John, in which Jesus raised Lazarus from the dead. Lazarus taxa are observational artefacts that appear to occur either because of (local) extinction, later resupplied, or as a sampling artefact. If the extinction is conclusively found to be total (global or worldwide) and the supplanting species is not a look-alike (an Elvis species), the observational artefact is overcome. The fossil record is inherently imperfect (only a very small fraction of organisms become fossilized) and contains gaps not necessarily caused by extinction, particularly when the number of individuals in a taxon becomes very low. If these gaps are filled by new fossil discoveries, a taxon will no longer be classified as a Lazarus taxon. A subtle difference is sometimes made between a "living fossil" and a "Lazarus taxon". A Lazarus taxon is a taxon (either one species or a group of species) that suddenly reappears, either in the fossil record or in nature, while a living fossil is a species that (seemingly) hasn't changed during its very long lifetime. Sometimes however, the two are confused or conflated, as with the coelacanth, which is also called a "living fossil" because it was thought to be extinct for tens of millions of years, but then discovered alive. (modified from Wikipedia)
Length: The length, or number of steps, is the total number of character state changes necessary to explain the relationship of the taxa in a tree. According to the principle of parsimony, the fewer number of character state changes required, the more likely the tree. A tree with a lower length has less homoplasies and so fits the data better than a tree with a higher length. The tree with the lowest length assumes fewer homoplasies and hence is more parsimonious, and so represents the hypothesis of taxa relationship that is selected. Lipscom 1998
Linnaean classification: hierarchical taxonomy developed by the 18th century Swedish botanist Carl von Linné, (Linnaeus). It was the first systematic classification of life on Earth, in which every species is given it's own binomial designation. So for example anatomically modern human beings are Homo sapiens, genus (the "family name") Homo and species (the specific name) sapiens. In contrast, Neanderthal man is Homo neanderthalensis. Linnaean classification provides a nested hierarchy of levels, each with its own specific characteristics. In this way any organism or species is grouped more and more specifically within the hierarchy. The Linnaean system was originally static, being based on creationism. In the 19th century, applied to the evolution of life and the modern synthesis it became evolutionary systematics, and was used to construct phylogenetic trees. Still foundational to modern biology, Linnaean classification is in the process of being superseded by phylogeny-based cladistic systematics. Unfortunately, this latter, with its indefinite series of nested clades, lacks the categorical simplicity and ease of use of the old Linnaean system. Some attempts have been made to integrate the two, but the incompatible methodologies mean these have not been very successful. More
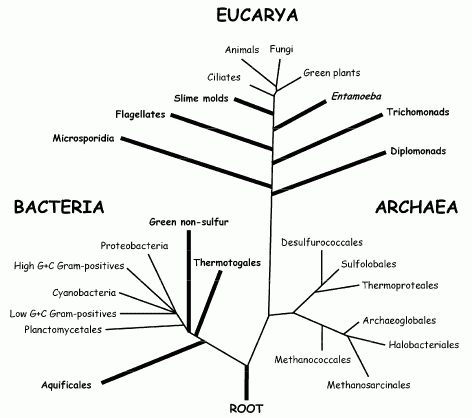 Diagram from Gribaldo & Philippe 2002. "The classical view of the universal tree of life, topology inspired from Stetter 1996, mainly based on rRNA comparison. Branches that could be affected by long branch attraction artefacts (e.g., the placement of the root in the bacterial branch or the early emergence of hyperthermophilic taxa amongst bacteria) are given as thick lines." |
Long branch attraction (LBA): A phenomenon in molecular phylogenetic analyses, especially those employing maximum parsimony. Unrelated species or lineages sharing rapid evolutionary rates are artefactually grouped together and hence considered closely related, regardless of their true evolutionary relationships. In other words, unrelated lineages may group on the basis of convergent changes rather than homologies, the long branches being attracted to each other because of chance similarities. For example, in DNA sequence-based analyses, the problem arises when sequences from two (or more) lineages evolve rapidly. For example, rRNA evolutionary rates may vary by a factor of 100 among planktonic foraminifers. As there are only four possible nucleotides, when DNA substitution rates are high, the probability that two lineages will evolve the same nucleotide at the same site increases. When this happens, parsimony erroneously interprets this homoplasy as a synapomorphy (i.e., evolving once in the common ancestor of the two lineages). In phylogenies rooted by a distant outgroup, unrelated fast evolving ingroups will emerge independently as the deepest offshoots, being attracted by the long branch of the outgroup. LBA artefact currently represents a major concern to phylogeneticists, as it is believed to affect the position of virtually every deep-branching lineage. As a result, many organismal relationships in the universal tree, shown as bold lines in the diagram on the right, should be regarded as suspect (note: this particular topology has since been corrected by more recent revisions) This problem can be minimized through improved models of sequence evolution and by using methods that correct for multiple substitutions at the same site, through increased or modified taxonomic sampling and by breaking up long branches adding taxa related to those with the long branches or by using alternative slower evolving traits. Long branche attraction is also a problem with morphology-based cladistics because each branch may have so many unique modifications that tracing shared (ancestral) conditions may be difficult. (Wikipedia, Gribaldo & Philippe 2002, Delsuc et al 2005, Edgecombe 2009) )
LUCA (Last Universal Common Ancestor): Also Universal Common Ancestor. The postulated most recent common ancestor of every living thing on Earth; the root of the tree of life. According to Carl Woese, horizontal gene transfer between the three domains early in the history of life makes the idea of a single common ancestor meaningless. More
Majority rule consensus: in cladistic analysis, a consensus method that preserves all relationships appearing in 50% of the source trees. This method allows a group to appear in the consensus even if some of the trees in the set contradict it, as long as a majority of the trees (generally half or more) support the grouping. In fully resolved majority rule consensus, these can appear in the consensus solution so long as they do not contradict relationships that occur more frequently. When comparing only two trees, this method is equivalent to the strict consensus method. (Bininda-Emonds, 2004 - glossary, from the PAUPDISPLAY Manual)
Matrix: tabulated data of the characters of all of the taxa used in a cladistic analysis, arranged in rows (taxon) and columns (character). "0" indicates that a character is absent, "1" that it is present. If there are more than one possible character states, these are inicated by further numbers, such as 2 or 3(very rarely more). if the character state is not known (common in the case of fossils, epsceially fragmentary ones), a question mark is used instead. The tabulated data is used to form phylogenetic hypotheses, which can be diagrammatically represented as cladograms. MAK120227
 Data matrix form the fossil lobster genus Hoploparia, from Tshudy & Sorhannus 2003 |
Maximum likelihood: In cladistics, one of several criteria that may be optimised in building phylogenetic trees from molecular sequence data. The maximum-likelihood method is a seemingly more powerful (and computationally intensive) parametric statistical technique than maximum parsimony, that uses an explicit model for character evolution and therefore is not subject to the same pitfalls of homoplasy and long branch attraction. Maximum likelihood will pick the most probable tree that explains the observed data. The optimal tree is the one that maximises the statistical likelihood that the specified evolutionary model produced the observed character-state data; the models specify the probabilities of character-state changes through evolutionary time. Compare with distance, parsimony. (Michael D. Crisp - Introductory glossary of cladistic terms; Nobu Tamura - Paleoexhibit)
Maximum parsimony: see parsimony
Molecular clock: the premise that the rate at which mutational changes accumulate is constant over time. The difference between the form of a molecules in two species is then assumed to be proportional to the time since the species diverged from a common ancestor, and molecules can be used to date the tree of life. In the late 1960s, the neutral theory of molecular evolution provided a theoretical basis for the molecular clock, though both the clock and the neutral theory were controversial, since most evolutionary biologists held strongly to panselectionism (Adaptationism), with natural selection as the only important cause of evolutionary change. (Wikipedia, etc). Although subject to certain caveats and continuing debate, the notion of the molecular clock has proven to be an important and useful tool in many contexts Searls, 2003 glossary The tendency now is to calibrate the molecular clock by the fossil record (Donoghue & Benton 2007). Earlier problems associated with this method for example, the evolution of animal phyla during the Precambrian (early in the Proterozoic (ref), for which there is absolutely no fossil evidence) have since been largely rectified. Even so, it is difficult to believe that the molecular clock rate does not vary greatly at particular times, for example accelerating during periods of rapid evolutionary radiation (the Cambrian explosion in this example). In other instances evolution may be more constant, and molecular clocks more reliable. The choice of molecule used may also be significant (reference to be included). (MAK)
Molecular phylogeny, Molecular systematics: Use of molecular data ((DNA, RNA, and/or proteins) as characters for phylogenetic analyses. That is, the use of the structure of molecules to gain information on an organism's evolutionary relationships. Includes methods based on overall similarity (Phenetics), like electrophoresis, immuno-distance and DNA-DNA-hybridisation, as well as methods that are based on parsimony, like restriction-site-analysis and sequencing sequencing). Generally speaking, the more closely related two organisms are, the more similar their gene sequences will be. By statistically comparing the similarities and differences in the sequence between the same gene from various organisms, we can deduce the pattern of how those organisms are related, and shown in a phylogram. Despite the similarities (both involve dichotomous branched trees), these are not cladograms! Over the last decade or so, molecular phylogeny has supplanted cladistic morphological phylogeny as the primary way of understanding the evolution of life on Earth, giving rise to phylogenetics, the synthesis of molecular phylogeny and phylogenetic systematics, based on a total evidence approach and supermatrix trees. (MAK) More.
Molecules: short for molecular sequencing or molecular phylogeny, and hence any resulting phylogenetic trees that may be derived from this methodology. One of the two rival phylogenetic methods currently in use, the other being morphology. Although molecular phylogeny has become the default paradigm (e.g. phylogenetics), here at Palaeos we have also given equal weight to morphology-based approaches MAK120326
Monophyletic, Monophyletic group, Monophyly: Originally coined by Haeckel to refer to a group of organisms that is descended from its most recent known or inferred common ancestor (Haeckel, 1866). A monophyletic group in this traditional sense of the word may include all or only a part of the descendants of the common ancestor, and the ancestor may be a taxon of various ranks (Mayr & Ashlock, 1991, Grant, 2003). Hennig (1966) restricts "Monophyly" to the only those groups in which no descendant is a part of any other group. Here monophyletic refers to a group containing all the inferred descendants of a common ancestor. Ashlock 1971 1974 proposed replacing Hennig's redefinition of monophyletic with the neologism holophyletic, but this suggestion has not widely caught on, and Hennig's terminology remains the most popularily accepted and indeed currently standard usage in the the scientific community. (MAK) More
Monotypic: in Linnaean classification, a higher rank taxon that contains only a single species. e.g. Ginkgo is a monotypic genus that contains a single extant species, biloba; the family Ginkgoaceae is similarily a monotypic family. In cladistics (and especially vertebrate paleontology), allowing only the type species in that genus; all other species are given their own genera. This is in keeping with a phylocode approach (which rejects supra-specific taxa such as genera, families, phyla etc), and understandable especially when dealing with fossil taxa where there is only very limited information (sometimes all that is known of a species are a few scraps of bone) and phylogenetic placement is uncertain.
Morphology: [1] The gross form and structure of an organism, or of a part of an organism. in paleontology and phylogeny may refer to the form or structure of a particular bone or shell, and its comparison with that of similar species. [b] Short for Morphology-based phylogeny (see next entry), and also referring to any resulting phylogenetic trees that may be derived from this methodology. One of the two rival phylogenetic methods currently in use , the other being molecular phylogeny. Although the latter has become the default paradigm (e.g. phylogenetics), here at Palaeos we have also given equal weight to morphology-based approaches. Morphology-based phylogeny is more or less synonymous with traditional cladistics, newer total evidence cladistic approaches also incorporate molecular sequencing data although the results may sometime sbe a little strange (such as pleurodire turtles as highly derived crown group cryptodires) MAK120326
Morphology-based phylogeny: Infrequently used term (and mostly by molecular phylogenists) to refer to, yes, you guessed it, phylogeny based on morphology rather than molecules. Synonymous for traditional cladistics
Neighbor-joining: a bottom-up clustering method for the creation of phenetic trees (phenograms), created by Naruya Saitou and Masatoshi Nei. Usually used for trees based on DNA or protein sequence data, the algorithm requires knowledge of the distance between each pair of taxa (e.g., species or sequences) in the tree. (Wikipedia

From Wikipedia. This genetic distance map made in 2002 is an estimate of 18 world human groups by a neighbour-joining method based on 23 kinds of genetic information. Public domain diagram by Jason Spatola.
Non- As cladistics does not allow the use of paraphyletic or ancestral taxa, it becomes difficult to refer to groups at the base of any evolutionary lineage. One way is to use prefixes like basal and stem, but these can tend to fuzzy vagueness, e.g. "basal archosauria" is not a correct term for all "thecodonts" but only strictly speaking refers to the most basal node or taxon of clade Archosauria. In this context, stem would be more accurate, but seems to be less often used. Another method is to use non-. For example, because the monophyletic clade Dinosauria includes not just dinosaurs but birds (because, cladistically speaking, birds are dinosaurs) dinosaurs as traditionally defined are not called dinosaurs but non-avian dinosaurs. The ancestors of dinosaurs, such as lagosuchids and silesaurids, then become non-dinosaurian dinosauromorphs. This problem does not arise in evolutionary systematics, which recognises and identifies ancestral groups. MAK120326
Numerical cladism: see Phylogenetic systematics.
Numerical taxonomy: same as phenetics; a method of generating phylogenies that is based on large numbers of quantifiable (measurable) characters which groups organisms with respect to overall similarity. (UCMP)
Node: any point in a cladogram where branches diverge or end. In cladistics, Nodes of phylogenetic trees represent taxonomic units. Internal nodes (or branches) refer to hypothetical ancestors whereas terminal nodes (or leaves).External nodes, which are at the end of a each branch represent terminal taxa, generally extant species but where paleontological data is considered they can also include fossil species. Internal nodes are where a single ancestral lineage breaks into two or more descendant lineages. In rooted trees, internal nodes represent hypothetical common ancestors. (Modified from Douglas Theobald's Phylogenetics Primer and UCMP Understanding Evolution Glossary)
Node-based taxon (or clade): in cladistics and phylogenetic nomenclature, all descendants of the most recent common ancestor of two or more specified taxa. A phylogenetically based taxon that does not require determining the presence or absense of apomorphies. Generally defined as "The least inclusive clade that includes taxon A + taxon B". Compare with stem-based taxon. MAK120318
Order: In the Linnaean classification the taxonomic rank between class and family, used to define middle-level sub-group. Orders are used in evolutionary systematics but not cladistics. (MAK) More
Overall similarity: method by which organisms that share the most similarities are grouped together; characters are not distinguished as to whether they are primitive or derived or whether they are evolutionary meaningful; also see numerical taxonomy (phenetics); contrast with phylogenetic systematics. (UCMP)
Outgroup: in cladistics, a taxon that is not part of the clade under consideration, but is including in the analysis in order to provide a baseline. In cladograms, outgroups are shown branching off at the base of the tree. (MAK)
Paleontology: the study of ancient life, on thebasis of fossil or other remains. More
Pan-group, Total group: A crown group and its stem group considered together. The Pan-Aves thus contain the living birds and all (fossil) organisms more closely related to birds than to crocodiles (their closest living relatives). Pan-Mammalia are all mammals and their fossil ancestors down to the phylogenetic split from the remaining amniotes (the Sauropsida). Pan-Mammalia is thus an alternative name for the clade Synapsida. With the exception of a few taxa, such as turtles, the pan-group approach has not caught on because it results in unnecessary junior synonyms. (Wikipedia)
Paraphyly, Paraphyletic group: neologism coined by Hennig (Hennig 1966) to refer to groups that have a common ancestry but that do not include all descendants (Horandl & Stuessy 2010, p.1642). They constitute one of the two types of monophyletic groups sensu Haeckel ; e.g. protist, reptile (see that entry for diagram), thecodont, condylarth; i.e. an ancestral taxon or evolutionary grade. Constitute "a group of organisms that has descended from a common ancestor but that does not include all descendants from this ancestor. A paraphyletic group of species was holophyletic before a younger derivative species (or derivatives) arose from that group" (Horandl & Stuessy 2010, p.1643). Cladists consider paraphyletic groups invalid (see e.g. Paraphyly watch blog), whereas evolutionary systematics regard them as perfectly acceptable. (MAK) More
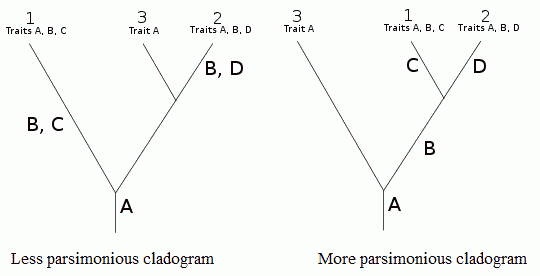 Use of parsimony in cladistics. It is considered more likely that trait B evolved only once (right hand cladogram) rather than twice (left-hand cladogram). Diagram adapted from Wikipedia. |
Parsimony: Also known as Occam's Razor (after the medieval theologian William of Ockham (c. 1285-1349), who rejected the idea of universals) is the principle that recommends when choosing between two competing hypotheses, that the simplest explanation of the evidence or observation is to be preferred, when the hypotheses are equal in other respects. A central premise in cladistics, where computer algorithms routinely generate huge numbers of cladistic trees. When reconstructing the phylogenetic relationships of a group of species or taxa, the principle of parsimony implies that we should prefer the the branching pattern or phylogeny that requires the fewest number of evolutionary changes (see above diagram). (MAK) Under maximum parsimony, the preferred phylogenetic tree is the one that requires the least number of evolutionary changes to explain the observed sets of characters (or traits). Whereas generally valid, this assumption can be problematic in cases such as homoplasy, when some of the traits are evolving much faster than others, or when some taxa have very long branches. Unfortunately, the picture becomes more complex when is taken into account. Compare Maximum likelihood. (Nobu Tamura - Paleoexhibit)
Pattern cladism, Transformed cladism: Dissenting Cladistic school, distinguished from phylogenetic or process cladism. Sometimes known as Cladists with a capital C (Williams and Ebach 2006). Transformed cladism is usually included here as well, although following Ebach et al 2008 they are given a separate entry. Founded by Gareth Nelson and Nelson Platnick ("New York Cladists") (Glossary of Phylogenetic Systematics - Günter Bechly although the latter is also associated with transformed cladism. (Ebach et al 2008).
As with phenetics, character rooting and synapomorphies are not used, although monophyletic groups are acknowledged. Essentially a reaction to Mayr's evolutionary systematics, Pattern Cladism constructs cladograms the pattern of their characters alone, without any recourse to evolution, through separation of "pattern and process". Unlike phenetics, pattern cladism distinguishes between synapomorphy, homoplasy and symplesiomorphy. Like phenetics, pattern and transformed cladists strove to be as objective and empirical as possible. Pattern Cladism asserts that a cladogram was merely a summary of shared characters, that could at best test a historical reconstruction (The philosophy of classification Pattern cladism and the myth of theory dependence of observation - John Wilkins) Even so, the idea that science can be theory-neutral is itself philosophically problematic (Pearson 2010). Criticised by Richard Dawkins' The Blind Watchmaker, (quotes and comments here, but see this defense of pattern and transform cladism. Ebach et al 2008 relate Transformed cladism to the works of Colin Patterson (1982) and Platnick (1979) and say that in contrast to pattern cladists who are Non-Hennigian, Transformed cladists are Hennigian and root their trees according to either outgroups, ontogeny or concepts such as set theory.... They opt for a definition of monophyly that does not include the most recent ancestor. They do not reject totally transformation, but they do use a concept of character rooting that is inherent within set theory". It seems however that these differences are mostly minor and the two are more usually synonymised. (MAK, IAB blog)
Phenetic species concept: A definition of a species as a set of organisms that are phenotypically similar to one another. Compare with biological species concept, cladistic species concept, ecological species concept, and recognition species concept. See other species definitions. (Fossil Mall glossary)
Phenetics, Phenetic systematics: School of numerical taxonomy developed in the late 1950s by bacteriologist Peter H. Sneath, entomologist Charles D. Michener, and quantitative geneticist Robert R. Sokal, that classifies organisms on the basis of overall morphological or genetic similarity. This mainly involves observable similarities and differences irrespective of whether or not the organisms are related. It involves grouping types together in clusters; types with many close relatives would be in a cluster. The development of Phenetics, which was intended to replace evolutionary systematics, was inspired through the quantitative successes and advances in genetics (e.g. discovery of DNA by Watson & Crick (1953)), chemistry and physics, on the other hand as a reaction to positivism and incorporation of a strictly pragmatic approach, which denies that we can know the thing in itself (hence the rejection of phylogeny and evolution). The availability of computers (at this time still big hulking things) also facilitated and encouraged quantitative data comparisons. This approach classifies organisms on overall similarity, usually in morphology or other observable traits, regardless of their phylogeny or evolutionary relation. It stressed the use of many unweighted characters assessed by overall similarity, purging all intuition and subjectivity and striving to be theory neutral, objective, and quantitative, with observation, description and ordering performed as precisely, objectively and repeatably as possible. Hence all evolutionary and phylogenetic interpretations are rejected as too difficult and subjective. It was considered that phylogenetic reconstruction was nearly impossible to know with any degree of certainty, and therefore, if classification were to be scientific, this futile quest should be abandoned. (Stuessy 2009, UCMP)
In the end Phenetics was unsuccessful and eventually abandoned in favour of cladistics for a number of reasons, including numerous difficulties encountered owing to convergence (homoplasy, as individual characters assumed to be homologous were not carefully analysed), mosaic evolution, and a shortage of diagnostic characters (Mayr & Ashlock 1991, pp. 195-205). Even so, certain phenetic methods, such as neighbor-joining, have found their way into cladistics, as a reasonable approximation of phylogeny when more advanced methods (such as Bayesian inference) are too computationally expensive. (Wikipedia) Also, with the rise of molecular systematics, distance methods, which are basically phenetic methods, have become popular, although these are vulnerable to the same problems, especially that of homoplasy. (Mayr & Bock 2002 p.180).
Phenetic pattern analysis: very similar to Phenetics and generally synonymised, it uses numerical methods for taxonomic classification.
Phenogram: A branching diagram (tree) showing the phenetic similarity among terminal taxa. Compare cladogram, dendrogram, phylogram. (Michael D. Crisp - Introductory glossary of cladistic terms)
Phenotype: The set of measurable or detectable physical or behavioral features of an individual. The phenotype represents the expression of the genotype of the individual as modified by environmental conditions during the individual's ontogeny. (W. R. Elsberry - talk.origins)
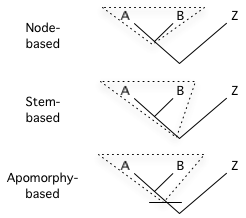
Phylocode: abbreviation for the International Code of Phylogenetic Nomenclature, a developing draft for a formal set of rules governing phylogenetic nomenclature. Its current version is specifically designed to regulate the naming of clades, leaving the governance of species names up to the rank-based codes. Unlike Linnaean-based nomenclatural codes the Phylocode does not require the use of ranks, although it does optionally allow their use. Rather than define taxa using a rank (such as genus, family, etc.) and a type specimen or type subtaxon, the content of taxa are delimited using a definition that is based on phylogeny (i.e., ancestry and descent) and uses specifiers (e.g., species, specimens, apomorphies) to indicate actual organisms. The formula of the definition indicates an ancestor. The defined taxon, then, is that ancestor and all of its descendants. Thus, the content of a phylogenetically-defined taxon relies on a phylogenetic hypothesis. In the Phylocode, clades may be node-based, stem-based, or apomorphy-based (see diagram at right).
The theoretical foundation of the Phylocode was developed in a series of papers by de Queiroz and Gauthier, which was foreshadowed by earlier suggestions that a taxon name could be defined by reference to a part of a phylogenetic tree. The number of supporters for official adoption of the Phylocode is still small, and it is uncertain, as of 2011, whether the code will be implemented and if so, how widely it will be followed. (Wikipedia)
Phylogenetic bracketing: see Extant Phylogenetic Bracket.
Phylogenetic hypothesis: an empirical hypothesis regarding evolutionary relationships suggested through cladistic or other phylogenetic methods. A phylogenetic hypothesis does not purport to describe the actual course of evolution itself, complete with ancestor descendent relationships, but rather a stylised or abstract representation of this (usually in the form of a cladogram or similar), based on available data, with the proviso that this can and indeed is likely to change or even be radically revised with new data, discoveries, and analyses. (MAK120318)
Phylogenetic incongruence: when two equally persuasive, verified, robust, and empirically supported methodologies give contrary phylogenetic results. For example, using morphology, the soft shelled turtles (Trionychia) are the most derived group of cryptodires, whereas using molocules, they are the most basal group. The problem here is in deciding which, if any, of the two methodologies provides the more reliable phylogenetic signal MAK120326
Phylogenetic nomenclature (or classification, or taxonomy): classification and taxonomy based on cladistic (Phylogenetic systematic) principles ("vertical" ancestry, not "horizontal" similarity), proposed as a rank-free alternative to the Linnaean system of classification, redefining taxa previously named under evolutionary systematics (e.g. Synapsida), and accepting only monophyletic clades. The goal is to make classification synonymous with phylogeny; i.e. to get rid of similarity altogether. Phylogenetic nomenclature has led to a number of controversial proposals, such as the abandonment of Linnaean binomial nomenclature, the rejection Linnaean ranks, and the migration of established names to crown clades (Benton 2007, p.651); e.g. Tetrapoda (this last reflecting an emphasis on neontology over paleontology that is still found in cladistics). Despite the logical and theoretical appeal of this approach, there are still problems in applying it in practice (Carlson, 2001, p.1113). See also Phylocode. (MAK)
Phylogenetic signal: the amount of information, or "signal" that can be retrieved from the background "noise" of any phylogenetic analysis or methodlogy. It is only to be expected that the advocates of any particular methodological paradigm consider that their own methodology provides the clearest phylogenetic signal. Therefore, in the case of any phylogenetic incongruency between themselves and a rival methodology, their own paradigm is automatically to be preferred. Take the example of molecular phylogeny verses cladistic morphology. Morphology strongly supports a monophyletic insectivora, based on a large number of unique shared characteristics, whereas molecular sequencing divides the insectivores into two unrelated clades, placing them in groups for which there is no morphological support. Because molecular phylogeny has replaced cladistics as the default option for any analysis that includes extant (recent) taxa, unqualified support of phylogenies resulting from this methodology, despite still being problematic are the standard approach. The unspoken implication here is that molecular phylogney has a much higher and more reliable phylogenetic signal, and that morphology involves so many convergences and reversals as to make extracting any possible phylogfenetic signal almost impossible, without firts being grounde din the molecular tree. As to why molecular sequencing, which is based largely on a phenetic approach, is considered superior is never explained, nor has its purported superioty been tested empirically. of course, it would be equally naive to accept the automatic superiory of morphology over molecules as well. Hence at Palaeos we have tried to adopt a non-partisan approach incorporating all methodolgies, popular and unpoular, the only proviso being that be scientific, verifable, and found in earlier or recent scientific literature. MAK120326
Phylogenetic systematics: Also known as Hennigian systematics, Numerical cladism, Phylogenetic cladism, and Process cladism, or sometimes just cladistics or phylogeny. Derives from Hennig's work and that of others such as James S. Farris, Walter Fitchand, and Herb Wagner. States that only shared derived characters can provide information about phylogeny . Those taxa that share a greater number of shared features are considered more closely related than those that don't. However, the shared (called characteristics have to be advanced (derived) rather than on primitive. The relationship between them is shown in a branching hierarchical tree called a cladogram. The cladogram is based on the principle that the fewest number of changes to map all the changes of character states is the most likely one; called the principle of parsimony. Only monophyletic groups are recognised. Unlike pattern cladism , which only aims at the calculation of most parsimonious cladograms from large data-sets, phylogenetic systematics also seeks to reconstruct phylogenetic schemes, in which all branching points are convincingly supported by characters, and using optimization (transformation series) (sensu Farris 1983) to select from a number of possible trees. (W. R. Elsberry talk.origins via W.J. Hudson; IAB blog, quoting Ebach et al 2008, Lipscom 1998, Günter Bechly, UCMP Understanding Evolution Glossary, and Wikipedia) More
Phylogenetic tree: See Tree.
Phylogenetics: A term derived from the Greek φῦλον, phulon, "tribe", γενέτης genetēs "ancestor" and -ικός, -ikos, an adjective-forming suffix. (Perseus Digital Library, Wiktionary) The synthesis of cladistics and molecular phylogeny, phylogenetics is the study of evolutionary relatedness among groups of organisms (e.g. species, populations), which is discovered through molecular sequencing data and morphological data matrices. (Wikipedia). See also Daniel F. Simola - Molecular Evolution and Phylogeny (pdf) for synoptic overview. More recently, phylogenetics has begun to incorporate other fields such as evo-devo as well (Telford & Budd 2003) which implies this is all leading to a new evolutionary synthesis (replacing the 20th modern synthesis).
Phylogenomics: the intersection between the fields of evolution and genomics. The use of cladistic principles to interpret genome data, and better understanding of gene function. One branch of phylogenomics involves the use of these data to reconstruct the evolutionary history of organisms. Another, "Pharmacophylogenomics" is the use of phylogenomics in aid of drug discovery, through improved target selection and validation. (Delsuc et al 2005, Searls, 2003, Wikipedia)
Phylogeny: term coined by Haeckel (Haeckel 1866): the study of the family history of life, the evolutionary relationships among groups of organisms, often illustrated with a branching diagram called a tree. There are several different forms:
Other approaches are possible too, for example developmental. In this way, phylogeny is used to understand the evolutionary history of life on Earth.
More colloqually or informally, phylogeny is also shorthand for any phylogenetic hypothesis or phylogenetic tree (e.g. cladogram). (MAK)
Phylogeography: research field that investigates the principles and processes that govern the geographic distributions of genealogical lineages, especially those within and among closely related species. (Phylogeography)
Phylogram: A phylogenetic tree usually distinguished from a cladogram in that the branch lengths are proportional to the amount of inferred evolutionary change. (Michael D. Crisp - Introductory glossary of cladistic terms, Wikipedia)
Phylo-optimism: the generally unspoken belief or premise held by some gradists that the fossil record, while admittedly incomplete, is still detailed enough to construct a reliable phylogeny or understanding of the path evolution actually took, as well as the characteristics of ancestral taxa and their descendants. Contrast with phylopessimism (MAK120318)
Phylopessimism: the generally unspoken belief or premise held by some cladists that the fossil record is so incomplete that we can never reconstruct a reliable or objective phylogeny; the best we can do is determine the most viable phylogenetic hypothesis using the incomplete data available to us. are not concerned by stratigraphic congruence and emphasise ghost lineages rather than look at alternative topologies. Contrast with phylo-optimism (MAK120318)
Phylum: In the Linnaean classification the taxonomic rank between kingdom and class, and hence one of the highest levels of taxonomic classification, used to define major groups of organisms; e.g. molluscs, arthropods, echinoderms, chordates. Phyla can be thought of as groupings of animals based on a shared general body plan. What this means is that despite the seemingly different external appearances of organisms, they can be classified into phyla based on their internal and developmental organizations. Despite their obvious differences, spiders and barnacles both belong to the phylum Arthropoda; but earthworms and tapeworms, although similar in shape, belong to different phyla. Although Linnaean rankings are not used in cladistic analysis, the majority of phyla are still accepted as they constitute monophyletic clades. (There are a few exceptions; e.g. growing consensus on the basis of molecular phylogeny is that Porifera (sponges) constitute an evolutionary grade. The rank of Phylum was not in Linneaus' original classification system, but was coined later by Haeckel. (MAK, Wikipedia)

Plesiomorphy, Plesiomorphic trait: in cladistic analysis, an ancestral or primitive character state present before the last common ancestor of the species group evolved, and hence not unique to the clade in question. Also called a primitive trait.
"Features shared more widely than in a group of interest. These are primitive for the group in question and cannot provide evidence for the group. An evolutionary trait that is homologous within a particular group of organisms but is not unique to members of that group (compare apomorphy) and therefore cannot be used as a diagnostic or defining character for the group. For example, vertebrae are found in zebras, cheetahs, and orangutans, but the common ancestor in which this trait first evolved is so distant that the trait is shared by many other animals. Therefore, possession of vertebrae sheds no light on the phylogenetic relations of these three species."
A Dictionary of Biology, Oxford University Press, © Market House Books Ltd 2000
Polarity: in phylogenetic cladistics this refers to the ordering of a particular character state, determined either independently of tree construction (direct method) or more usually from a rooted tree (indirect method) (Michael D. Crisp - Introductory glossary of cladistic terms) To quote Telford & Budd 2003 p.487: "In order for an analysis to be useful in an evolutionary sense, it needs to be rooted, in other words we need to know the polarity of change of the characters that interest us. If we consider two taxa in isolation (say a lizard and a mouse) that differ in a certain character (e.g. hairless or hairy) how do we know which of the two has the primitive character state and which the derived? ...(T)o determine the direction in which the evolution of this character has proceeded...knowledge of the state of the character in a species that is an outgroup (is needed)...in this case, a frog would be appropriate. As the frog is hairless, parsimony suggests that hairlessness is the primitive character and we can infer from this that hair has evolved in the lineage leading to mice after this lineage had diverged from reptiles." Polarity is one of the ways in which phylogenetic systematics is distinguished from non-phylogenetic ordering systems such as phenetics and pattern cladistics.
Polyphyly, Polyphyletic group: A group that does not share a common ancestor, but is defined on the basis of independently acquired or convergent (non-homologous) character states. Examples for polyphyletic groups would be the old taxon Pachydermata which includes the thick-skinned hippos, rhinos and elephants, or the taxon Haemothermia (endorsed at one time by Lovtrup and Gardiner) for a grouping of haemothermic birds and mammals. Polyphyletic groups are considered invalid by both evolutionary and phylogenetic systematics. (MAK, Glossary of Phylogenetic Systematics - Günter Bechly)
Polytomy: in a cladistic phylogeny, a node where more than two lineages descend from a single ancestral lineage. This indicates either that we don't know how the descendant lineages are related or the descendant lineages speciated simultaneously. Where a branching pattern cannot be resolved, the branches in question can be collapsed to show the absence of a hypothesis for the relationships among the lineages that they represent. (UCMP Understanding Evolution Glossary; Keeling & Palmer 2008 p.607 ).
Primitive trait: same as plesiomorphy; a character that is present in the common ancestor of a clade; a primitive trait is inferred to be the original character state of that character within the clade under consideration; compare to derived trait. (UCMP)
Process cladism: see Phylogenetic Systematics.
Pseudomonophyletic: neologism coined by MAK for an artefact of cladistic methodology (whether morphological or molecular or both) in which a taxon that appears to be a robust monophyletic clade in initial analyses, but is later shown to be a paraphyletic or polyphyletic taxon; e.g. Ceratosauria, Nyctiphruretia, Cyclostomata. Pseudomonophyletic taxa would seme to be the result of widespread reversals among ancestral (basal, stem, plesiomorphic, non-, select your preferred prefix) taxa. The loss of shared character traits among these primitive forms, and hence the absense of what should be plesiomorphic (shared primitive) traits in more derived taxa gives the appearance that the ancestral grade is a monophyletic clade. MAK120326
Rank: the hierarchical level of a supra-specific taxon, according to the Linnaean approach to classification. The eight ranks are kingdom, phylum (added by Haeckel), class, order, family, tribe (used mostly in botany, much more rarely in zoology and paleontology), genus, and species, plus optional intermediate grades represented by the suffixes super-, sub- and infra-. ( In the three domain theory of Carl Woese and co-workers, a further rank, domain, is sometimes added above kingdom, although it seems to me that domain and kingdom are just different ways of approaching the same topic (like evolutionary and phylogenetic systematics)). The highest ranks are the most general, whilst each sub-division or rank adds it own increasingly specific and unique characteristics. In this way any organism or species can be grouped more and more specifically within the hierarchy. A central part of evolutionary systematics, according to which every taxon that evolves from another taxon has the same taxonomic rank. So class Reptilia would give rise to class Mammalia, not subclass Mammalia (although, Reptilia includes a number of subclasses, these are still part of the class Reptilia). Determining the appropriate rank is an art, not a mechanical process, and inevitably ranks don't always equate. e.g. the orders of modern birds are probably equivalent to families or superfamilies of fish or invertebrates. Ranks are strenuously rejected by most cladists, although some paleontologists such as Michael Benton argues that cladistics and Linnaean ranks are not be incompatible. Moreover, ranks are useful in everything from field guides (compare the muddled organisation of taxa in Greg Paul's otherwise superlative Princeton Field Guide to Dinosaurs with the clear arrangement of Predatory Dinosaurs of the World from more than two decades earlier) to measuring the degree of biodiversity through time. (MAK)
Relationship: the way two or more species or other taxa are evolutionarily and phylogenetically related on an evolutionary tree. In cladistics, two taxa may have a sister relationship, or one may be more basal or more derived than the other. In evolutionary systematics, one may be the ancestor of another, or they may share an actual common ancestor. (MAK120318)
Retention index (RI): Similar to the consistency index, but defined so that the highest possible value for any character is 1.0 and the lowest is 0.0; removes bias due to autapomorphies. (Michael D. Crisp - Introductory glossary of cladistic terms). The per-character retention index (ri) is defined as (g-s)/(g-m), where where m is the minimum possible number of character changes (steps) on any tree, s the actual number of steps on the current tree, and g is the maximal number of steps for the character on any cladogram (Farris 1989). The retention index measures the amount of synapomorphy on the tree, and varies from 0 to 1. (Øyvind Hammer - PAST - Paleontological Statistics Software)
Reversal: in cladistics, the loss of a character trait, or more technically, the evolutionary reversion from an apomorphic to a plesiomorphic character state. Compare with homoplasy.
Robust: a phylogenetic hypothesis or tree topology that is supported by a large number of character states and/or molecular sequencing results, and not destabilised through the addition of one or two phylogenetic steps hence can be considered reliable. MAK120326
Romerogram: A bubble diagram or spindle diagram that plots diversity (horizontal axis, width of bubble or spindle) against time (vertical axis), and showing the phylogenetic divergence of new groups from ancestral lineages. The changing width of the bubbles represents the increase or decrease in diversity or abundance of individuals and species of a particular taxon through time. Named after the great vertebrate paleontologist Alfred Sherwood Romer, who popularised the use of such diagrams. Disliked by cladists because of their use of paraphyletic taxa, although Michael Benton (Benton 2004) uses a modified version that emphasises monophyletic clades. (MAK111018)
Root: in cladistics, the common ancestor of all taxa represented in a cladogram. The root is often determined using an outgroup taxon to determine the evolution in the taxa of interest (Delsuc et al 2005). See also base node.
Rooted tree: A cladogram with a hypothetical ancestor, which equates to the root. When outgroups are used, this is the node that connects the outgroups to the ingroup, and which thus specifies the direction of evolutionary change among the character-states. Contrast with unrooted tree. (Michael D. Crisp - Introductory glossary of cladistic terms)
Scala Naturae: a Latin expression meaning "natural ladder", is a sort of proto-taxonomy first developed by Aristotle, according to which the natural world can be arranged in a single linear series from inanimate matter through plants, invertebrates, higher vertebrates, and finally man. Along with Plato's Principle of Plenitude it led to the idea of the Great chain of being. Scala Naturae and Great Chain of Being remained central ideas in natural philosophy until the mid 19th century. More
Semistrict consensus: also called "combinable component" consensus. If a particular grouping in one tree is not contradicted by the other trees, it will be retained in the consensus. When there is a conflict in grouping, semistrict consensus behaves like strict consensus. (from the PAUPDISPLAY Manual)
Sequencing: any of several methods and technologies that are used for determining the order of proteins in a cell, or nucleotide bases (adenine, guanine, cytosine, and thymine) in a molecule of RNA or DNA. An essential element in modern biological systematics (molecular phylogeny). The rapid speed of sequencing attained with modern DNA sequencing technology has been instrumental in the sequencing of the human genome (the Human Genome Project). Related projects, often by scientific collaboration across continents, have generated the complete sequences of many animal, plant, and microbial genomes. (Wikipedia)
Similarity: the degree to which two or more species resemble or don't resemble each other is one of the two factors that could be considered in any biological classification and taxonomy, the other being phylogeny. Similarity could be the result of common descent and divergence (homology) or convergence (homoplasy). Pre-Darwinian natural philosophy considered only similarity (being unaware of phylogeny), evolutionary systematics gave equal weight to both, phenetics and pattern cladistics rejected phylogeny as impractical and thus revert to similarity only, whilst Hennigian cladistics and phylogenetic nomenclature goes to the other extreme and rejects similarity altogether, emphasising only phylogeny. (MAK)
Singly paraphyletic group: a group or taxon that is paraphyletic because one of its descendant lineages are not included. e.g. Class Amphibia is a singly paraphyletic group because amphibians evolved into reptiles. There are also doubly, triply and so on paraphyletic groups (MAK120318)
Sister group: Cladistic term for any of the descendant branches from a node on a cladogram. In a phylogeny, the descendants of an ancestor are called daughters, while the siblings after a speciation event are called sisters (so a descendant is a daughter relative to its ancestor and is a sister relative to its other sibling). Note that if either of the daughters undergoes further speciation then the sister to a particular terminal taxon may actually be a group of terminal taxa. (Michael D. Crisp - Introductory glossary of cladistic terms)
Species: Highly controversial term given a variety of definitions by biologists. Currently, the Biological Species Concept (BSC) is widely popular: Groups of actually or potentially interbreeding populations, which are reproductively isolated from other such groups (Mayr 1963) . (W. R. Elsberry talk.origins via W.J. Hudson) See also cladistic species concept, ecological species concept, phenetic species concept, and recognition species concept. (Fossil Mall glossary) More
 The spindle diagram shown on the right is typical of mid 20th century phylogenies. This particular diagram which I found through Google image search is from an online creationist book ( original url). The taxa correspond to orders, subclasses, or classes, in the Linnaean ranking. The caption reads: Fig. 20. Family tree of the vertebrates. On the left is a geological time scale. Phylogenetic origins of various groups of vertebrates. A) Urodela, B) Lepospondyli, C) Apoda, D) Anura, E) Labyrinthodontia, F) Apsidospondyli, G) Chelonia, H) Anapsida, I) Cotylosauria, J) Euryapsida, K) Diapsida, L) Eosuchia, M) Squamata, N) Rhynchocephalia, O) Ornithischia, P)Thecodontia, Q) Synapsida, R) Parapsida, S) Pelycosauria, T) Pterosauria, U) Crocodilia, V) Aves, W) Saurischia, X) Prototheria, Y) Metatheria, Z) Pantotheria, AA) Therapsida, BB) Eutheria, CC) Ichthyosauria. The width of the spindle shows taxonomic diversity, numeric abundance (the abundance of fossils of the group in strata of the particular geologic period), or both; the distinction here being often poorly defined. In an effort to introduce precision, spindle width may be drawn according to the number of genera or families known from a particular time period, but even determining what qualifies as a genus or family can be arbitrary (see splitters versus lumpers). Contrast this diagram with cladistic dendrograms and cladograms which show relationships between individual species, without referencing time, transformation, or evolutionary lineage. (MAK) |
Spindle diagram: An evolution tree that maps lineage diversity or abundance mapped against geologic time. Spindle diagrams are employed in evolutionary taxonomy, but not phylogenetic systematics. They frequently emphasise ancestral or paraphyletic groups, transitional forms, and transformation of one lineage into another. Note the way the various more recent spindles emerge from earlier lineages. Also called a Bubble diagram or Romerogram.
Stem: ancestral forms, species or taxa that constitute the trunk of an evolutionary tree rather than the later ramifications.
Stem-based taxon (or clade, or group): in cladistics and phylogenetic nomenclature, all species, living or extinct, that share a more recent common ancestor (or Last Common Ancestor - LCA) with a specified species than with other specied species or taxa. When only two species are refferd to it may be abbreviated to Anchor Taxon > Another taxon. > is the mathematical symbol for "greater than", in this case it means more similar to. A Stem-based group is a phylogenetically based taxon that does not require determining the presence or absense of apomorphies. Compare with node-based taxon. MAK120318
Stem group: Not to be confused with stem-based group (see above entry), the concept of stem groupsis used in cladistics to cover evolutionary "aunts" and "cousins" of living groups. A crown group is a group of closely-related living animals plus their last common ancestor plus all its descendants. A stem group is a set of offshoots from the lineage at a point earlier than the last common ancestor of the crown group; it is a relative concept, for example tardigrades are living animals which form a crown group in their own right, but Budd 1996 and 2001 regarded them also as being a stem group relative to the arthropods. Stem group shown in yellow in this diagram. (Wikipedia). The distinction however between the stem- and crown group is an arbitrary one, because it is determined only by the most basal member of the crown group that is still extant; if Branchiostoma had gone extinct in the Pleistocene, or even the 15th century, our concept of the euchordate crown-group be radically different, because Branchiostoma lacks many features of higher chordates (Budd 2001 p.265).
Because cladistics has replaced evolutionary sytematics, but does not include all of the concepts provided by the latter, there is a tendency for terms like stem and basal to appear as rather vague alternatives to "primitive" or "ancestral" in cladistic paleontological literature and especially popularised accounts and comments thereof. An alternative is the use of the prefix non-. MAK120318 120417
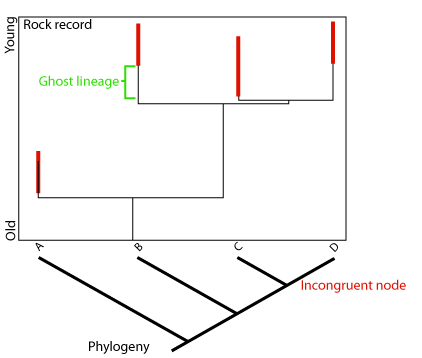 How do we identify ghost lineages and measure their prevalence in a cladogram. All other things being equal, we expect the terminal taxa that branch off of a cladogram first to appear first in the fossil record. When this is true, the cladogram is said to be stratigraphically congruent. Often, cladograms are not stratigraphically congruent. This happens when there are long ghost lineages. - diagram and caption by John Merck. |
Stratigraphic congruence: The degree to which the terminal taxa that branch off of a cladogram match the order with which they first appear in the fossil record. A simple measure of stratigraphic congruence is the Stratigraphic Congruence Index (SCI) of Huelsenbeck (1994) is defined as the proportion of stratigraphically consistent nodes on the cladogram, and varies from 0 to 1. A node is stratigraphically consistent when the oldest first occurrence above the node is the same age or younger than the first occurrence of its sister taxon (node). (Øyvind Hammer - PAST - Paleontological Statistics Software)
Stratigraphy: in the phylogenetic context, the chronological and stratigraphic order by which taxa appear in the fossil record, in this context refers to biostratigraphy, rather than stratigraphy as such. Here, the more primitive or ancestral form should always preceed the more advanced type of organism. So, for example, the protodinosaurs ("non-dinosaurian dinosauromorpha" to use the unwieldly cladistic definition) appear in the middle Triassic period, whereas their descendents the true dinosaurs only appear some ten or twenty million years later in the late Triassic. However the vagaries of the fiossil record mean that not every evolutionary lineage is recorded, for this reason most cladists, apart from the rarely used stratocladistic approach, ignore stratigraphic sequence and fill in the gaps with ghost lineages. MAK120326
Stratocladistics: a method of making phylogenetic inferences using both geological and morphobiological data. It is similar to cladistics in emphasising parsimony and synapomorphies, but also incorporating stratigraphic information as well; in this way temporal data are used along with conventional character data and Bayesian logic to selecting most parsimonious hypotheses. (Fisher, 2008)
Stratophenetics: phylogenetic method based on (a) the identification of taxa based on phenetic similarities among specimens and (b) stratigraphic interval, so that taxa from different time-intervals are linked in presumed ancestor-descendant sequences according to their similarities. - (from Michal Kowalewski, lecture notes)

Strict consensus: this is the most conservative consensus method used in in cladistic analysis, which only recognises clades that appear in all of the trees. It's advantage is that it only includes data that is totally unambiguous. The disadvantage is that it is thrown off by the slightest difference. For example, two trees may be identical except for the placement of a single sequence, yet their strict consensus tree might be completely unresolved. The resulting is a "star" phylogeny, a broad polytomy with only radiating lines, and very little or no resolution or phylogenetic structure. This is shown by the blue cladogram on the right, which is placed next to a more conventional, branching phylogeny. (from the PAUPDISPLAY Manual, Forey et al 1992 p.78)
Supermatrix: One of the new developments in cladistics that have become possible through cheap and powerful computing, supermatrixes involve simultaneous analysis of all available character data. Rather than separate analyses of data sets and subsequent integration of the resulting trees (supertree), all character data is considered simultaneously to enable incorporation of diverse kinds of data, including characters from fossils, morphology, and molecular phylogeny. (de Queiroz & Gatesy 2007) (MAK)

Supertree: In cladistics, a "supertree" refers to the synthesis of a number of distinct cladograms, combining morphological, molecular, and other data from the different individual phylogenies. Supertrees result from combining many smaller, overlapping phylogenetic trees into a single, more comprehensive tree. They are distinguished from classic
consensus techniques in that the source trees need only have overlapping rather than identical taxon sets. Because supertree construction uses other tree topologies rather than the primary data underlying those trees, they can be constructed using all available phylogenetic hypotheses, even those based on incompatible data types, or lacking data entirely. Supertree have produced phylogenies of a number of large taxonomic groups. However supertree strength is also its weakness, and this approach has been harshly criticised by systematists precisely because it only considers the topology of the source trees, effectively discarding primary data. A supertree or quasi-supertree approach is also standard with ASCII phylogenetic trees. Supertree construction is probably as old as the field of systematics itself, and remains our only way of visualizing the Tree of Life as a whole. References: Pisani et al 2002 who give the example of a dinosaur supertree (see diagram, right), Bininda-Emonds, 2004. (MAK)
Supra-specific taxon: a taxon above the species level: anything from subgenus and genus upwards (family, order, etc). Useful for understanding biotic diversity through time and large scale patterns of evolution. Recognised by evolutionary systematics, but not by cladistics. See also rank. (MAK)
Symplesiomorphy A shared plesiomorphic character trait, which is shared between two or more taxa, but which is also shared with other taxa which have an earlier last common ancestor with the taxa under consideration. An example is pharyngeal gill breathing in bony and cartilaginous fishes. The former are more closely related to Tetrapoda (terrestrial vertebrates, which evolved out of a clade of bony fishes) that breathe via their skin or lungs, rather than to the sharks, rays, etc. Their kind of gill respiration is shared by the "fishes" because it was present in their common ancestor and lost in the other living vertebrates. Contrast with apomorphy/synapomorphy. (Wikipedia)
Synapomorphy: an apomorphy that is shared by (syn-) by several taxa, where the trait in question originates in their last common ancestor. Being shared by multiple taxa, synapomorphies can be used to diagnose (describe) a clade (a monophyletic group). Compare with homology. True synapomorphies usually are a given set of terminal groups, shared by two or more terminal taxa, but this is not essential to the concept. Thus, if some descendants of a last common ancestor possess a synapomorphic trait, in the case of reversals it is not strictly necessary that all of its descendants must possess the same trait. Contrast with plesiomorphy and homoplasy, which are shared primitive and shared convergent characteristics also of no phylogenetic value. (based on Wikipedia, Michael D. Crisp - Introductory glossary of cladistic terms)
Systematics, Systematic biology: as defined here, the study of the diversification of life on the planet Earth, both past and present, and the relationships among living things through time. Relationships are visualized as evolutionary trees. Here there are two main paradigms, evolutionary systematics (now rarely used in veretbrate paleontology) and phylogenetic (cladistic) systematics. Evolutionary systematics interprets Linnaean classification in terms of the modern evolutionary synthesis and provides evolutionary taxonomies above the species level. It maps lineages against a geological time to give a spindle diagram showing diversity or abundance. Cladistic phylogenies (cladograms) are based at the species level and emphasise greater verifiability. They have two components, branching order (showing group relationships) and (in the case of phylograms) branch length (showing amount of evolution). Almost all systematics nowadays is cladistically-derived. Phylogenetic trees of species and higher taxa are used to study the evolution of traits (e.g., anatomical or molecular characteristics) and the distribution of organisms (biogeography).
Systematic biology, taxonomy, and scientific classification are often confused and used interchangeably. However, taxonomy is more specifically the identification, description, and naming (i.e. nomenclature) of organisms, classification focuses on placing organisms within hierarchical groups that show their relationships to other organisms, and systematics alone deals specifically with relationships through time, and can be synonymous with phylogenetics, broadly dealing with the inferred evolutionary hierarchy of organisms. (MAK, Wikipedia)
Systematic paleontology: organising or classifying fossil organisms (paleontology) according to the principles of systematic biology (Systematics). (MAK)
Taxon: (plural: taxa) a group of organisms, considered to be a unit, and which generally has been formally named with a scientific (Latin or Greek) proper name and a rank. Defining what belongs or does not belong to such a taxonomic group is done by a taxonomist with the science of taxonomy. It is not uncommon for one taxonomist to disagree with another on what exactly belongs to a taxon, or on what exact criteria should be used for inclusion. Traditionally, a taxon is given a formal or scientific name, which is governed by one of the Nomenclature Codes, which sets out rules to determine which scientific name is correct for that particular grouping. Generally, a good taxon as one that reflects presumptive evolutionary (phylogenetic) relationships, being derived from a common ancestor . Whether or not clades are acceptable as taxons is a matter of dispute; although evolutionary systematists (Mayr & Bock 2002 p.182) deny that they are, whereas cladists have proposed phylogenetic nomenclature and a new Phylocode which requires taxa to be monophyletic and rejects Linnaean supra-specific ranks. (MAK, Wikipedia)
Taxonomy: the practice and science of classification. Taxonomy uses taxonomic units, known as taxa (singular taxon). In addition, the word is also used as a count noun: a taxonomy, or taxonomic scheme, is a particular classification ("the taxonomy of ..."), arranged in a hierarchical structure. See also Alpha taxonomy , cladistics, Linnaean classification, Systematics (Wikipedia). In biological taxonomy there are, according to Ereshefsky (2000, p. 7) "no fewer than four general schools of taxonomy: evolutionary taxonomy, pheneticism, process cladism, and pattern cladism". Each of those schools have their own view on how to get from the characteristics of an individual organism to a species, and also the meaning of the term species varies between schools of taxonomy. (cited from Birger Hjørland)
Tetrapod: four-legged, land-living vertebrate, or any secondarily limbless (e.g. snakes) or aquatic (e.g. whales) descendants of such. Cladistic terminology disagrees over whether "tetrapod" should be used to include all four-legged animals (stem-based definition) or only those that include the common ancestor of all living tetrapods and its descendants (crown-based definition). More
Terminal, Terminal taxon: Not the end of the evolutionary line, but in cladistic formalism, one of the units whose collective phylogeny is reconstructed; shown diagramatically as the undivided tips of a cladogram. Terminals may be higher taxa, species, populations, individuals, fossils or even genes. There should be some rational basis for accepting the integrity of each terminal (for the purpose of the analysis), e.g. a monophyletic or diagnosable unit. Despite the claims by some authors, terminals do not need to be monophyletic; in fact, many species-level terminals are unavoidably paraphyletic. However, higher taxa used as terminals should be monophyletic. (based on Michael D. Crisp - Introductory glossary of cladistic terms)
Three-domain system: biological classification introduced by Carl Woese that rejects the old prokaryote-eukaryote distinction and divides cellular life forms into Archaea, Bacteria, and Eukarya domains (usually interpreted as a taxonomic grade above kingdom). Woese argued that, on the basis of differences in 16S rRNA genes, the three groups each arose separately from an ancestor with poorly developed genetic machinery, called a progenote. To reflect these primary lines of descent, he treated each as a domain, divided into several different kingdoms. He conjectured an era in which there was a considerable amount of lateral transfer of genes between organisms. Species formed when organisms stopped treating genes from other organisms with equal importance to their own genes. Lateral transfer during this period was responsible for the fast early evolution of complex biological structures. (MAK, corrected from Wikipedia also Wikipedia). Refs Woese et al 1990
Topology: in this context, the particular shape or arrangement of the branches of a cladogram, the evolutionary history of the group in question according to a particular phylogenetic hypothesis. (MAK120318)
Total evidence: the philosophical principle that the best hypothesis is the one derived from all the available data. In cladistics, this principle has come to be equated with the supermatrix approach (Bininda-Emonds, 2004 - glossary).
Total group: see Pan-group.
Transformed cladism: see Pattern cladism.
Tree: also Phylogenetic tree: a branching tree-like, diagrammatic representation of the evolutionary relationships and patterns of branching in the history of the organisms being considered. One type of phylogenetic tree, called a cladogram, is central to cladistics (especially phylogenetic systematics). Dendrogram is sometimes used to refer to a more informal diagram. (MAK) More
Triply paraphyletic group: a group or taxon that is paraphyletic because three of its descendant lineages are not included. There would also be quadruply paraphyletic groups and so on. See also singly and doubly paraphyletic groups
Unrooted tree: A cladogram for which the ancestor (the root) has not been hypothesized, and which thus does not specify the direction of evolutionary change among the character-states. An unrooted tree can be rooted on any of its branches, and so there are many rooted trees that can be derived from a single unrooted tree. Contrast with rooted tree. (Michael D. Crisp - Introductory glossary of cladistic terms)
Vertical classification: as described by Simpson, a taxon based on ancestor and descendant (phylogenetic) relationship between its members, a clade. Evolutionary systematics considers both horizontal and vertical classification in taxonomy, whereas cladistics (phylogenetic systematics) is based on vertical classification only. (MAK) More
Vraagteken effect: from the Dutch "question mark", a term introduced by Schram and Hof (1998) to the effect that the absence of critical information has in destabilising cladograms. They found that by introducing fossil taxa, for which, obviously, many character states are unknown, and hence coded as a question mark in the matrix, resulted in a great variation in the topology of the trees recovered by parsimony analysis. Cladistic algorithms respond to the ambiguity caused by the missing data by generating a large number of equally parsimonious phylogenies MAK120417.
Wastebasket taxon: a taxon that includes all species or groups that cannot be easily or conveniently placed elsewhere, e.g., for a while all large theropod dinosaurs that could not be included under the Ceratosauridae, Allosauridae or Tyrannosauridae were named "Megalosaurus".
Weighting: in cladistics, the empirically controversial (because non-quantifiable) yet necessary task of determining the phylogenetic significance of a particular character trait. For example, if there are three species of animals, one with brown fur, another with black fur, and one with brown scales, the presence or absence of fur is more important than the external colour, and hence would be given greater weight in phylogenetic analysis. Weighting is unavoidable if one is to address the problem of homoplasy vs homology. (MAK)
| Page Back: Molecular phylogeny | Unit Home | Page Top | Page Next: References |
content by MAK110419, edited RFVS111203, last modified MAK120326
